
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,922,996 – 19,923,087 |
| Length | 91 |
| Max. P | 0.639126 |

| Location | 19,922,996 – 19,923,087 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.64 |
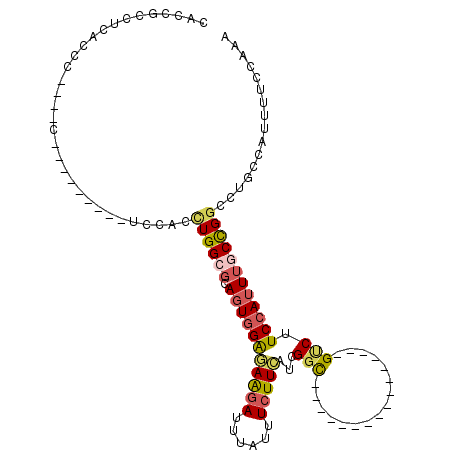
| Mean single sequence MFE | -22.30 |
| Consensus MFE | -11.00 |
| Energy contribution | -11.34 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
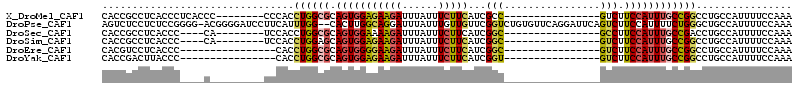
>X_DroMel_CAF1 19922996 91 - 22224390 CACCGCCUCACCCUCACCC--------CCCACCUGGCGCAGUGGAGAAGAUUUAUUUCUUCAUCGCC----------------GUCUUCCAUUUGCCGGCCUGCCAUUUUCCAAA ....(((............--------.......)))((((..(((((((......))))).))(((----------------(.(........).))))))))........... ( -20.61) >DroPse_CAF1 38479 112 - 1 AGUCUCCUCUCCGGGG-ACGGGGAUCCUUCAUUUGG--CACUUGGCAGGAUUUAUUUGUUGUUCGGUCUGUGUUCAGGAUUCAGUCUUCCAUUUUCUGGGCUGCCAUUUUCCAAA .......((((((...-.)))))).......(((((--....(((((((((..(........)..))))..((((((((..............)))))))))))))....))))) ( -31.24) >DroSec_CAF1 37139 87 - 1 CACCGCCUCACCC----CA--------UCCACCUGGCGCAGUGGAAAAGAUUUAUUUCUUCAUCGGC----------------GCCUUCCAUUUGCCGACCUGCCAUUUUCCAAA ...((((......----..--------.......))))...(((((((((......))....(((((----------------(.........)))))).......))))))).. ( -18.36) >DroSim_CAF1 28595 87 - 1 CACCGCCUCACCC----CA--------UCCACCUGGAGCAGUGGAGAAGAUUUAUUUCUUCAUCGGC----------------GUCUUCCAUUUGCCGGCCUGCCAUUUUCCAAA .............----..--------......(((((.(((((.(((((......))))).(((((----------------(.........))))))....)))))))))).. ( -22.80) >DroEre_CAF1 34150 83 - 1 CACGUCCUCACCC----------------CACCUGGCGCAGUGGGGAAGAUUUAUUUCUUCAUCGGC----------------GUCUUCCAUUUGCCGGCCUGCCAUUUUCCAAA .(((((.....((----------------(((........)))))(((((......)))))...)))----------------))............((....)).......... ( -20.20) >DroYak_CAF1 35621 83 - 1 CACCGACUUACCC----------------CACCUGGCGCAGUGGAGAAGAUUUAUUUCUUCAUCGGU----------------GUCUUCCAUUUGCCGGCCUGCCAUUUUCCAAA ((((((......(----------------(((........)))).(((((......))))).)))))----------------).............((....)).......... ( -20.60) >consensus CACCGCCUCACCC____C_________UCCACCUGGCGCAGUGGAGAAGAUUUAUUUCUUCAUCGGC________________GUCUUCCAUUUGCCGGCCUGCCAUUUUCCAAA ................................((((((.(((((((((((......)))))...(((................))).))))))))))))................ (-11.00 = -11.34 + 0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:14 2006