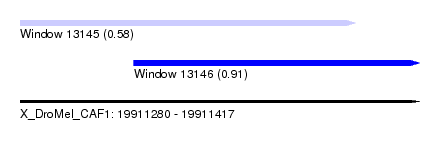
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,911,280 – 19,911,417 |
| Length | 137 |
| Max. P | 0.908358 |

| Location | 19,911,280 – 19,911,395 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.86 |
| Mean single sequence MFE | -40.90 |
| Consensus MFE | -21.89 |
| Energy contribution | -22.12 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19911280 115 + 22224390 GUGCUCUCGAUUGCAAAUCCCG-GCCAUAAUUUAUGCCUAUGGC-CG-GAGUCUGCAUUGAUAUUUUGAUGCAAAGCAGUUGGGUUGAACUGAGCUGACAGACAGCUGGCACAGGA--CU ((((((((((((((.....(((-((((((.........))))))-))-)....(((((..(....)..)))))..)))))))))........(((((.....))))))))))....--.. ( -47.50) >DroPse_CAF1 26955 116 + 1 CCUCUCUCGAUGCCAAAUCCCGGGCCGUAAUUUAUGCCUAUGGCCCG-AAGUACGCAUUGAUAUUUUGAUGCAAAGCAGUUGAGUUAA---CAACUGACAAACAGCUGGCACGAGCAGCU ...((((((.((((......(((((((((.........)))))))))-.(((..((((..(....)..))))...(((((((......---)))))).).....))))))))))).)).. ( -42.30) >DroEre_CAF1 23064 116 + 1 GUGCUCUCGAUUGCAAAUCCCG-GCCAUAAUUUAUGCCUAUGGCCCG-GAGUGUGCAUUGAUAUUUUGAUGCAAAGCAGUUGGGUUCAACUGAGCUGACAGACAGCUGGCAUGGGA--CU ((((((((((((((...(((.(-((((((.........))))))).)-))...(((((..(....)..)))))..)))))))))........(((((.....))))))))))....--.. ( -44.10) >DroYak_CAF1 24915 116 + 1 GUGCUCUCGAUUGCAAAUCCCG-GCUAUAAUUUAUGCCUAUGGCCCG-GAGUGUGCAUUGAUAUUUUGAUGCAAAGCAGUUGGGCCGAACUGAGCUGACAGACAGCUGGCACAGGA--CU ((((((((((((((...(((.(-((((((.........))))))).)-))...(((((..(....)..)))))..)))))))))........(((((.....))))))))))....--.. ( -43.90) >DroAna_CAF1 47546 111 + 1 ---CACUCGAUUGCAAAUCCUGGGCUGUAAUUUAUGCCUAUGACCCAAAAGUAUGCAUUGAUAUUUUGAUGCAAAGCAGUUGAGUUAA---CAACUGACAGGCAGCUGGCACC-GA--CU ---.((((((((((...((.(((((..........))))).))..........(((((..(....)..)))))..))))))))))...---((.(((.....))).)).....-..--.. ( -31.40) >DroPer_CAF1 30227 116 + 1 CCUCUCUCGAUGCCACAUCCCAGGCCGUAAUUUAUGCCUAUGGCCCA-AAGUACGCAUUGAUAUUUUGAUGCAAAGCAGUUGAGUUAA---CAACUGACAAACAGCUGGCACGAGCAGCU ...((((((.((((........(((((((.........)))))))..-.(((..((((..(....)..))))...(((((((......---)))))).).....))))))))))).)).. ( -36.20) >consensus GUGCUCUCGAUUGCAAAUCCCG_GCCAUAAUUUAUGCCUAUGGCCCG_AAGUAUGCAUUGAUAUUUUGAUGCAAAGCAGUUGAGUUAA___CAACUGACAGACAGCUGGCACGGGA__CU .....(((((((((......((.((((((.........)))))).)).......((((..(....)..))))...)))))))))..........(((.....)))............... (-21.89 = -22.12 + 0.22)



| Location | 19,911,319 – 19,911,417 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 71.31 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -18.52 |
| Energy contribution | -18.48 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19911319 98 + 22224390 UGGC-CG-GAGUCUGCAUUGAUAUUUUGAUGCAAAGCAGUUGGGUUGAACUGAGCUGACAGACAGCUGGCACAGGACUCAGAGGACUUGGAAGAC------------------UCGGA ...(-((-.(((((((((..(....)..)))).((((..(((((((...((((((((.....)))))....))))))))))..).)))...))))------------------)))). ( -31.70) >DroSec_CAF1 25454 90 + 1 UGGCCCG-GAGUCUGCAUUGAUAUUUUGAUGCAAAGCAGUUGGGUUGAACUGAGCUGACAGACAGCUGGCACAGGACUC---------GGAAGAC------------------UCGGA (((((.(-..((((((((..(....)..))))...((((((.((.....)).))))).)))))..).))).)).((.((---------....)).------------------))... ( -28.50) >DroEre_CAF1 23103 108 + 1 UGGCCCG-GAGUGUGCAUUGAUAUUUUGAUGCAAAGCAGUUGGGUUCAACUGAGCUGACAGACAGCUGGCAUGGGACUCAGAGGACUGG---------GAGGACUGGGCGGACUGGGA ..(((((-(.((((((((..(....)..))))....((((((....))))))(((((.....))))).))))....(((((....))))---------)....))))))......... ( -38.00) >DroYak_CAF1 24954 117 + 1 UGGCCCG-GAGUGUGCAUUGAUAUUUUGAUGCAAAGCAGUUGGGCCGAACUGAGCUGACAGACAGCUGGCACAGGACUCGGAGGACUCGAAGGACUCGGAGGACUCAGAGGACUGGAA .((((((-(....(((((..(....)..)))))......)))))))......(((((.....)))))....(((..(((.(((..(((((.....)).)))..))).)))..)))... ( -47.30) >DroAna_CAF1 47583 85 + 1 UGACCCAAAAGUAUGCAUUGAUAUUUUGAUGCAAAGCAGUUGAGUUAA---CAACUGACAGGCAGCUGGCACC-GACUCCAAGUCCUUU----------------------------- .(((......((.(((((..(....)..)))))...((((((......---)))))))).((.((.((....)-).))))..)))....----------------------------- ( -19.50) >consensus UGGCCCG_GAGUCUGCAUUGAUAUUUUGAUGCAAAGCAGUUGGGUUGAACUGAGCUGACAGACAGCUGGCACAGGACUCAGAGGACUUG_A_GAC__________________UCGGA .((((((...((.(((((..(....)..)))))..))...))))))......(((((.....)))))................................................... (-18.52 = -18.48 + -0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:09 2006