
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,907,232 – 19,907,375 |
| Length | 143 |
| Max. P | 0.990168 |

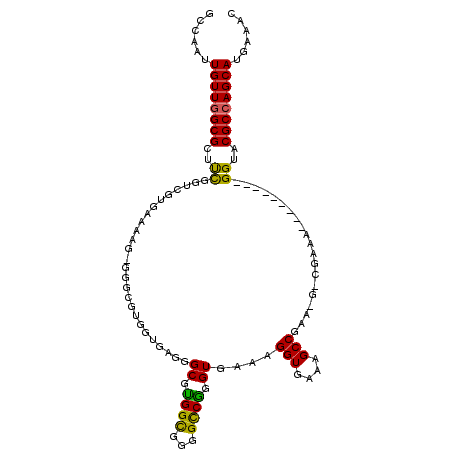
| Location | 19,907,232 – 19,907,336 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.45 |
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -22.56 |
| Energy contribution | -22.92 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19907232 104 + 22224390 GCCAAUUGUUGGCGCUUCGGUCGUGAAAGGGGGGCGUGGUGGGGGCGCGGCGAGGCCGGGUGAAAGGUGAAAGCCGAAAGGCGAAA---------GGUACGCCAGCAUGAAAC (((..(..((.((((....).))).))..)..)))(((.(((.(((.((((...)))).))...........(((....)))....---------....).))).)))..... ( -38.80) >DroSec_CAF1 21292 96 + 1 GCCAAUUGUUGGCGCUUCGGUCGUGAAAAG-GGGCGUGGUGAGGGCGUGGCGGGGCCGGGUGAAAGGUAAAAGCCGA-------AA---------GGUACGCCAGCAUGAAAC ((((.....))))(((.((.((.(....).-)).)).)))....((.(((((..(((........)))....(((..-------..---------))).)))))))....... ( -33.10) >DroSim_CAF1 17372 96 + 1 GCCAAUUGUUGGCGCUUCGGUCGUGAAAGG-GGGCGUGGUGAGGGCGUGGCAGGGCCGGGUGAAAGGUAAAAGCCGA-------AA---------GGUACGCCAGCAUGAAAC (((.....((.((((....).))).))...-.))).(.(((..(((((......(((........)))....(((..-------..---------))))))))..))).)... ( -33.10) >DroEre_CAF1 20388 91 + 1 GCCAAUUGUUGGCGCUCCUGGCGGG--AA-----------GGGGGCGUGGCGGGGUCAGGUGAAAGGUGAAAGCCGAAAGCCGAAA---------GGUGCGCCAGCAUGAAAG ......(((((((((.(((..(((.--..-----------....((.((((...)))).))....(((....))).....)))..)---------)).)))))))))...... ( -38.10) >DroYak_CAF1 21646 102 + 1 GCCAAUUGUUCGCGUUUUUGGGCGGAAAA-----------GAGGGCGUGGUGGGGCCAGGUGAAAGGUGAAAGCCGAAUGCCGAAAGGUACGAAAGGUACGCCAGCAUGAAAC (((..((.(((((.(....).))))).))-----------...))).(((((..(((........(((....)))...((((....)))).....))).)))))......... ( -34.90) >consensus GCCAAUUGUUGGCGCUUCGGUCGUGAAAAG_GGGCGUGGUGAGGGCGUGGCGGGGCCGGGUGAAAGGUGAAAGCCGAA_G_CGAAA_________GGUACGCCAGCAUGAAAC ......((((((((..((..........................((.((((...)))).))....(((....)))....................))..))))))))...... (-22.56 = -22.92 + 0.36)


| Location | 19,907,268 – 19,907,375 |
|---|---|
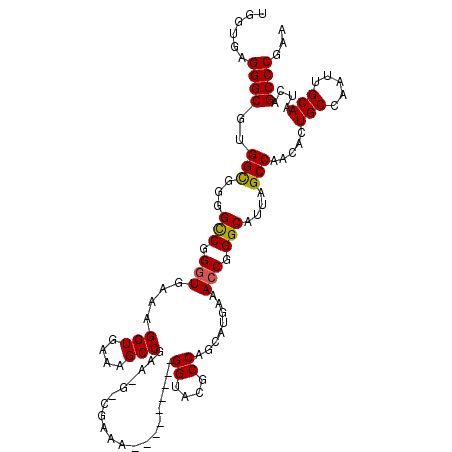
| Length | 107 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 85.02 |
| Mean single sequence MFE | -37.66 |
| Consensus MFE | -28.88 |
| Energy contribution | -29.96 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19907268 107 + 22224390 UGGUGGGGGCGCGGCGAGGCCGGGUGAAAGGUGAAAGCCGAAAGGCGAAA---------GGUACGCCAGCAUGAAACCGGGCAUUAGCCAACACUGCCAAUUGCAAUCAGCCCGAA ......((((..(((...(((.(((.....((....(((....)))....---------((....)).)).....))).)))....))).....(((.....)))....))))... ( -39.30) >DroSec_CAF1 21327 100 + 1 UGGUGAGGGCGUGGCGGGGCCGGGUGAAAGGUAAAAGCCGA-------AA---------GGUACGCCAGCAUGAAACCGGGCAUUAGCCAACACUGCCAAUUGCAAUCAGCCCGAA ......((((.((((...(((.(((....(((....(((..-------..---------)))..)))........))).)))....))))....(((.....)))....))))... ( -35.20) >DroSim_CAF1 17407 100 + 1 UGGUGAGGGCGUGGCAGGGCCGGGUGAAAGGUAAAAGCCGA-------AA---------GGUACGCCAGCAUGAAACCGGGCAUUAGCCAACACUGCCAAUUGCAAUCAGCCCGAA .(((((..((.((((((...((((((...(((....(((..-------..---------)))..)))..)))....)))(((....)))....))))))...))..))).)).... ( -35.10) >DroEre_CAF1 20415 102 + 1 ----GGGGGCGUGGCGGGGUCAGGUGAAAGGUGAAAGCCGAAAGCCGAAA---------GGUGCGCCAGCAUGAAAGCGGGCAUUAGCCAACACUGCCAGUUGCAAUCAGCCC-AA ----..((((.((((((.((..(((....(((....)))(...(((....---------))).)(((.((......)).)))....))).)).))))))(.......).))))-.. ( -38.40) >DroYak_CAF1 21675 112 + 1 ----GAGGGCGUGGUGGGGCCAGGUGAAAGGUGAAAGCCGAAUGCCGAAAGGUACGAAAGGUACGCCAGCAUGAAACCGGGCAUUAGCCUACACUGCCAAUUGCAAUCAGCCCGAA ----..(((((((...((((..((((...(((....)))...((((....))))(....)...)))).((.((....)).))....)))).)))(((.....)))....))))... ( -40.30) >consensus UGGUGAGGGCGUGGCGGGGCCGGGUGAAAGGUGAAAGCCGAA_G_CGAAA_________GGUACGCCAGCAUGAAACCGGGCAUUAGCCAACACUGCCAAUUGCAAUCAGCCCGAA ......((((..(((...(((.(((....(((....)))....................((....))........))).)))....))).....(((.....)))....))))... (-28.88 = -29.96 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:06 2006