
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,904,004 – 19,904,206 |
| Length | 202 |
| Max. P | 0.823474 |

| Location | 19,904,004 – 19,904,095 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 85.55 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -22.28 |
| Energy contribution | -22.68 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19904004 91 + 22224390 AUGUCACCUGCAUGCUGAACGAGCGUUUUGAUUGCGU------GUGUGUUCAGUGUUCA--------------GUGUACAUAGGUGCAUAAGUG--UGCGUGGCAUGUUGGAU ((((((((((.((((((((((.((((.......))))------...)))))))))).))--------------).((((((..........)))--))))))))))....... ( -32.20) >DroSec_CAF1 18325 97 + 1 AUGUCACCUGCAUGCUGAACGAGCGUUUUGAUUGCGUUGUGGCGUGUGCUCAGUGUUCA--------------AUGUACAUUGGUGCAUAAGUG--UGCGUGGCAUGUUGGAU ......((.((((((((((((((((.......(((((....))))))))))...)))).--------------(((((((((........))))--)))))))))))).)).. ( -31.61) >DroSim_CAF1 13639 96 + 1 AUGUCACCUGCAUGCUGAACGAGCGUUUUGAUUGCGUUGUGGCGUGUGCUCAGUGUUCA--------------GUGUACAU-AGUGCAUAAGUG--UGCGUGGCAUGUUGGAU (((((((..((((((((((((((((.......(((((....))))))))))...)))).--------------((((((..-.)))))).))))--))))))))))....... ( -29.81) >DroEre_CAF1 17274 101 + 1 AUGUCACCUGCA-GCUGAGCGAGCGUUUUGAUUGCGUUGUGGCAAGUGUACAGUGUACACUGUAC---------UGUACAUAGGUGCAUAAGUG--UGCGUGGCAUGUUGGAU .((((((.((((-((.((((....)))).).)))))..)))))).((((.((.((((((((....---------(((((....)))))..))))--))))))))))....... ( -31.60) >DroYak_CAF1 18250 113 + 1 AUGUCACCUGCAUGCUGAACGAGCGUUUUGAUUGCGUUGUGGCGUGUGCACAGUGUACACUGUACAGUGUACAGUGUACAUAGGUGCAUAAGUGUGUGCGUGGCAUGUUGGAU (((((((.((((((((..((.(((((.......)))))))....(((((((.(((((((((((((...)))))))))))))..))))))))))))))).)))))))....... ( -48.40) >consensus AUGUCACCUGCAUGCUGAACGAGCGUUUUGAUUGCGUUGUGGCGUGUGCUCAGUGUUCA______________GUGUACAUAGGUGCAUAAGUG__UGCGUGGCAUGUUGGAU ......((.(((((((((((....)))).....((((....((.(((((...((((.((...............)).))))....))))).))....))))))))))).)).. (-22.28 = -22.68 + 0.40)



| Location | 19,904,038 – 19,904,135 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.03 |
| Mean single sequence MFE | -24.45 |
| Consensus MFE | -15.32 |
| Energy contribution | -15.98 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19904038 97 - 22224390 GGGAAAUUAAUUUUGAUAUGUUGAUUUCUCGCAUGGGCAAAUCCAACAUGCCACGCA--CACUUAUGCACCUAUGUACAC--------------UGAACACUGAACACAC------ACG (((((((((((........)))))))))))(((((((((.........))))..(((--......)))...)))))....--------------................------... ( -20.00) >DroPse_CAF1 20855 79 - 1 GGGAAAUUAAUUUUGAUAUGUUGAUUUCUCGCAUGAGCAAAUCCAACAUGCCACACGGACACGG------------ACAC---------------GAACACC-------C------ACA (((..........((.((((((((((((((....))).)))).))))))).))..((....)).------------....---------------.....))-------)------... ( -17.00) >DroSec_CAF1 18359 103 - 1 GGGAAAUUAAUUUUGAUAUGUUGAUUUCUCGCAUGGGCAAAUCCAACAUGCCACGCA--CACUUAUGCACCAAUGUACAU--------------UGAACACUGAGCACACGCCACAACG (((((((((((........)))))))))))((.((((....))))...(((((.(((--......)))..((((....))--------------)).....)).)))...))....... ( -20.50) >DroSim_CAF1 13673 101 - 1 GGGAAAUUA-UUUUGAUAUGUUGAUUUCUCGCAUGGGCAAAUCCAACAUGCCACGCA--CACUUAUGCACU-AUGUACAC--------------UGAACACUGAGCACACGCCACAACG (((((((((-...........)))))))))(((((((((.........))))..(((--......)))..)-))))....--------------.......((.((....))))..... ( -18.00) >DroEre_CAF1 17307 108 - 1 GGGAAAUUAAUUUUGAUAUGUUGAUUUCUCGCAUGGGCACAUCCAACAUGCCACGCA--CACUUAUGCACCUAUGUACA---------GUACAGUGUACACUGUACACUUGCCACAACG (((((((((((........)))))))))))...((((((......(((((....(((--......)))...)))))...---------(((((((....)))))))...)))).))... ( -30.30) >DroYak_CAF1 18284 119 - 1 GGGAAAUUAAUUUUGAUAUGUUGAUUUCUCGCAUGGGCAAAUCCAACAUGCCACGCACACACUUAUGCACCUAUGUACACUGUACACUGUACAGUGUACACUGUGCACACGCCACAACG (((((((((((........)))))))))))((.((((((.........)))).............(((((...((((((((((((...))))))))))))..))))))).))....... ( -40.90) >consensus GGGAAAUUAAUUUUGAUAUGUUGAUUUCUCGCAUGGGCAAAUCCAACAUGCCACGCA__CACUUAUGCACCUAUGUACAC______________UGAACACUGAGCACACGCCACAACG (((((((((((........)))))))))))(((((...........)))))...(((........)))................................................... (-15.32 = -15.98 + 0.67)

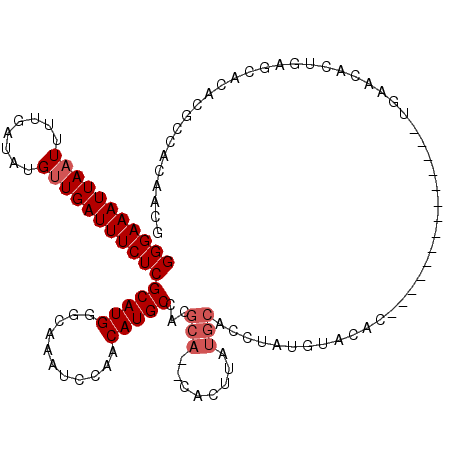

| Location | 19,904,095 – 19,904,206 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.71 |
| Mean single sequence MFE | -22.58 |
| Consensus MFE | -17.32 |
| Energy contribution | -17.07 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19904095 111 - 22224390 CCAAGGGUGUGAGCCCA-------CAUUCGCCACCCUCUUAUGCAUUAAAUUGACAUUCAUAUUACUUAUUUAUGCAUGGGAAAUUAAUUUUGAUAUGUUGAUUUCUCGCAUGGGCAA ...((((((((((....-------..)))).))))))(((((((.......((.(((..(((.....)))..))))).(((((((((((........))))))))))))))))))... ( -30.40) >DroPse_CAF1 20894 99 - 1 ------------GCCCU-------CCGCCGCCCCUCUUAUAUGCAUUAAAAUGACAUUCAUAUUACUUAUUUAUGCAUGGGAAAUUAAUUUUGAUAUGUUGAUUUCUCGCAUGAGCAA ------------.....-------.................(((..(((.(((.....))).))).....((((((..(((((((((((........)))))))))))))))))))). ( -19.10) >DroEre_CAF1 17375 94 - 1 ------------------------CAUCCGCCAUCCUGCUAUGCAUUGAAUUGACAUUCAUAUUACUUAUUUAUGCAUGGGAAAUUAAUUUUGAUAUGUUGAUUUCUCGCAUGGGCAC ------------------------.....(((....(((.(((((((((((....)))))............))))))(((((((((((........))))))))))))))..))).. ( -24.00) >DroYak_CAF1 18363 112 - 1 ------CAGCGACCUCCCAUUCGCCAUUCGCCACCCUCCUGUGCAUUAAAUUGACAUUCAUAUUACUUAUUUAUGCAUGGGAAAUUAAUUUUGAUAUGUUGAUUUCUCGCAUGGGCAA ------..((((........)))).............(((((((.......((.(((..(((.....)))..))))).(((((((((((........))))))))))))))))))... ( -25.30) >DroAna_CAF1 41839 96 - 1 -------------CCC--------CCAUCCCCA-CAUCGUAUGCAUUAAAUUGACAUUCAUAUUACUUAUUUAUGCAUGGGAAAUUAAUUUUGAUAUGUUGAUUUCUCGCAUGAGCAA -------------...--------.........-.......(((.......((.....))..........((((((..(((((((((((........)))))))))))))))))))). ( -17.60) >DroPer_CAF1 24086 99 - 1 ------------GCCCU-------CCUCCGCCCCUCUUAUAUGCAUUAAAAUGACAUUCAUAUUACUUAUUUAUGCAUGGGAAAUUAAUUUUGAUAUGUUGAUUUCUCGCAUGAGCAA ------------.....-------.................(((..(((.(((.....))).))).....((((((..(((((((((((........)))))))))))))))))))). ( -19.10) >consensus ____________GCCC________CAUCCGCCACCCUCAUAUGCAUUAAAUUGACAUUCAUAUUACUUAUUUAUGCAUGGGAAAUUAAUUUUGAUAUGUUGAUUUCUCGCAUGAGCAA .........................................(((.......((.....))..........((((((..(((((((((((........)))))))))))))))))))). (-17.32 = -17.07 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:02 2006