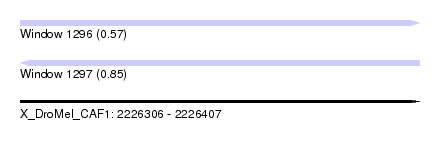
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,226,306 – 2,226,407 |
| Length | 101 |
| Max. P | 0.849278 |

| Location | 2,226,306 – 2,226,407 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.82 |
| Mean single sequence MFE | -35.22 |
| Consensus MFE | -22.67 |
| Energy contribution | -22.32 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2226306 101 + 22224390 CGAAGCGCUGGCGGCGUCGCCGACAGCCACCGAGCAGCCACUGUCAGCAGUAGCGCCAGAUUUUUUUCGAGGUAGCUCCUAAACAGA------------AACCUUCAUUAUUU .((((.((((.((((...)))).))))....((((.((.((((....)))).))(((.((......))..))).)))).........------------...))))....... ( -32.10) >DroGri_CAF1 2326 112 + 1 CGAAGCGUUGACGGCGUCGACGACAUCCGCCGAGUAGCCACUGUCAGCAGUAGCGCCAGAUUUUCUUUGAGGUGGCUGCUGCAAUCAAAUUUC-ACAAAAGUCUCGUUUAUUA ..(((((..((((((.(((.((.....)).)))...))).......(((((((((((.((......))..))).))))))))...........-......))).))))).... ( -33.90) >DroSec_CAF1 3959 109 + 1 CGAAGCGCUGGCGGCGUCGCCGACAACCACCGAGCAGCCACUGUCAGCAGUAGCGCCAGAUCUUUUUCGAGGUAGCUCCUAAACGGAAACAC----GGAAAUCUUCAUUAUUG .((((..((((((((...((((........)).)).)))((((....))))...)))))....(((((((((.....)))....(....).)----))))).))))....... ( -32.70) >DroEre_CAF1 4873 107 + 1 CGAAGCGCUGGCGGCGCCGCCGACAACCACCGAGCAGCCACUGUCAGCAGUAGCGCCAGAUCUUCUUCGAGGUGGCUCCUAAACAGAAAUUU----UCAGAAU--UUUCAUUU .((((.((((((((.((.((((........)).)).))..))))))))..(((.((((..((......))..))))..))).........))----)).....--........ ( -29.00) >DroYak_CAF1 5031 109 + 1 CGAAGCGCUGGCGGCGCCUCCGGCAGCCACCGAGCAGCCACUGUCAGCAGUAGCGCCAGAUCUUCUUCGAGGUGGCUCCUAAGCAGAAUAAU----GCAUUAUAUCAUCAAUU .((.((((.....)))).)).((.((((((((.((.((.((((....)))).))))).((......))..)))))))))...(((......)----))............... ( -36.80) >DroMoj_CAF1 2896 107 + 1 CGAAGCGCUGGCGGCGCCGGCGGCAGCCGCCGAACAGCCACUGUCAGCAGUAGCGCCAGAUUUUCUUUGAGGUGGCUGCUAAA------CACCGAAAAGAAGAUCGAUUAUUC ....((((((((((((((...))).))))))........((((....)))))))))..((((((((((..((((.........------))))..))))))))))........ ( -46.80) >consensus CGAAGCGCUGGCGGCGCCGCCGACAGCCACCGAGCAGCCACUGUCAGCAGUAGCGCCAGAUCUUCUUCGAGGUGGCUCCUAAACAGAAACAU____AAAAAACUUCAUUAUUU ......((((((((.((.((((........)).)).))..))))))))((.((((((.((......))..))).))).))................................. (-22.67 = -22.32 + -0.36)

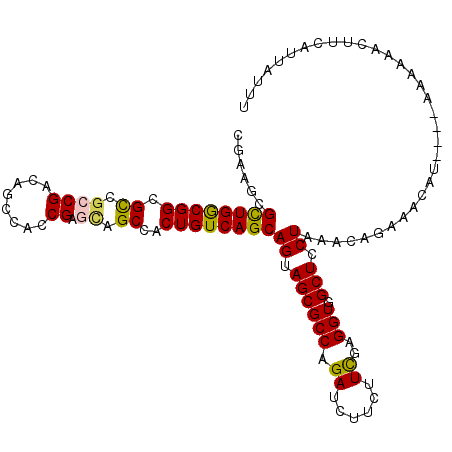
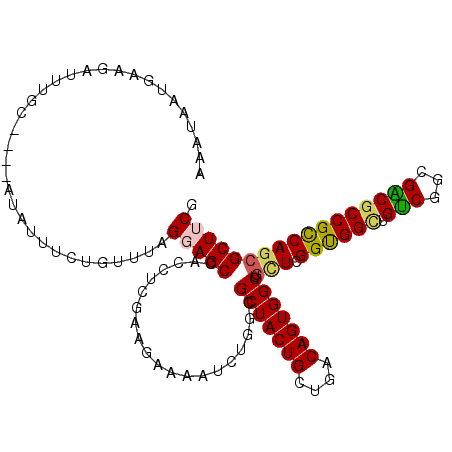
| Location | 2,226,306 – 2,226,407 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.82 |
| Mean single sequence MFE | -37.64 |
| Consensus MFE | -30.48 |
| Energy contribution | -30.20 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2226306 101 - 22224390 AAAUAAUGAAGGUU------------UCUGUUUAGGAGCUACCUCGAAAAAAAUCUGGCGCUACUGCUGACAGUGGCUGCUCGGUGGCUGUCGGCGACGCCGCCAGCGCUUCG .......(((((((------------(((....))))))...............((((((....((((((((((.(((....))).))))))))))....))))))..)))). ( -38.80) >DroGri_CAF1 2326 112 - 1 UAAUAAACGAGACUUUUGU-GAAAUUUGAUUGCAGCAGCCACCUCAAAGAAAAUCUGGCGCUACUGCUGACAGUGGCUACUCGGCGGAUGUCGUCGACGCCGUCAACGCUUCG .......((((.....((.-.(.......)..))((.((((..............))))(((((((....))))))).....(((((.((((...)))))))))...)))))) ( -29.04) >DroSec_CAF1 3959 109 - 1 CAAUAAUGAAGAUUUCC----GUGUUUCCGUUUAGGAGCUACCUCGAAAAAGAUCUGGCGCUACUGCUGACAGUGGCUGCUCGGUGGUUGUCGGCGACGCCGCCAGCGCUUCG .......((((.((((.----(((.((((.....)))).)))...)))).....((((((....(((((((((..(((....)))..)))))))))....))))))..)))). ( -36.20) >DroEre_CAF1 4873 107 - 1 AAAUGAAA--AUUCUGA----AAAUUUCUGUUUAGGAGCCACCUCGAAGAAGAUCUGGCGCUACUGCUGACAGUGGCUGCUCGGUGGUUGUCGGCGGCGCCGCCAGCGCUUCG ........--.((((((----(........)))))))...........((((..((((((...((((((((((..(((....)))..))))))))))...))))))..)))). ( -38.30) >DroYak_CAF1 5031 109 - 1 AAUUGAUGAUAUAAUGC----AUUAUUCUGCUUAGGAGCCACCUCGAAGAAGAUCUGGCGCUACUGCUGACAGUGGCUGCUCGGUGGCUGCCGGAGGCGCCGCCAGCGCUUCG ...............((----(......)))...(((((((((((......))...((((((((((....))))))).))).))))))).)).(((((((.....))))))). ( -41.80) >DroMoj_CAF1 2896 107 - 1 GAAUAAUCGAUCUUCUUUUCGGUG------UUUAGCAGCCACCUCAAAGAAAAUCUGGCGCUACUGCUGACAGUGGCUGUUCGGCGGCUGCCGCCGGCGCCGCCAGCGCUUCG ........(((.((((((..((((------.........))))..)))))).))).((((((((((....))))))).(((.((((((.(((...)))))))))))))))... ( -41.70) >consensus AAAUAAUGAAGAUUUGC____AUAUUUCUGUUUAGGAGCCACCUCGAAGAAAAUCUGGCGCUACUGCUGACAGUGGCUGCUCGGUGGCUGUCGGCGACGCCGCCAGCGCUUCG ..................................(((((....................(((((((....))))))).(((.((((((.(((...))))))))))))))))). (-30.48 = -30.20 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:07 2006