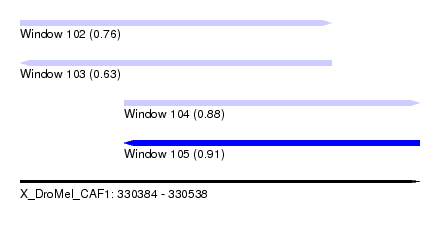
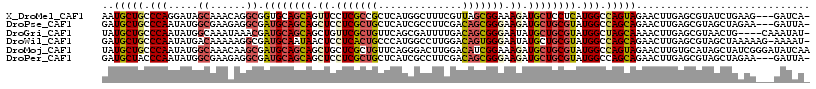
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 330,384 – 330,538 |
| Length | 154 |
| Max. P | 0.911853 |

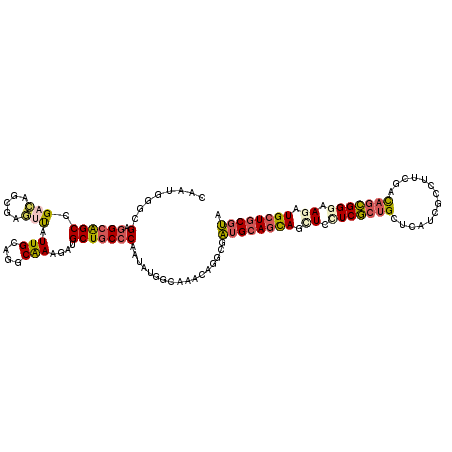
| Location | 330,384 – 330,504 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.28 |
| Mean single sequence MFE | -45.02 |
| Consensus MFE | -25.53 |
| Energy contribution | -25.92 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 330384 120 + 22224390 CAAUGGGGGAGGCAGCCGUUAAGGAUCCAUUGCAUGCGAAAAUGCUGCCCAGGAUAGCAAACAGGCGGUGCAGCAGUUCCUCGCCGCUCAUGGCUUUCGUUAGCGGAAAGAUGCUCCUCA .....((((((.((.(((((((.((.((((.((..((((...((((((((......((......)))).)))))).....)))).))..))))...)).))))))).....)))))))). ( -44.70) >DroPse_CAF1 1637 120 + 1 CAAUGGGCGAGGCGGCCGACAGCGAGUUAUUGAAGGCAAAGAUGCUGCCCAAUAUGGCGAAGAGGCGAUGCAGCAGCUCCUCGCUGCUCAUCGCCUUCGACAGCGGGAAGAUGCUGCGUA ...(((((..(((((((..(((.......)))..))).....))))))))).....(((..(((((((((.((((((.....)))))))))))))))...(((((......)))))))). ( -48.70) >DroGri_CAF1 39492 120 + 1 CAAUCGGCGAUGCAGCCGAUGGUGAAUUGUUGCCAGCAAAUAUGCUGCCCAAUAUGGCAAAUAAACGAUGCAGCAGCUGUUCGCUGUUCAGCGAUUUUGACAGCGGGAAUAUGCUGCGUA ..((((((......))))))((..(....)..))((((....))))(((......))).........((((((((....((((((((.(((.....)))))))))))....)))))))). ( -45.20) >DroWil_CAF1 1611 120 + 1 CAAUCGGCGAGGCGGCUGAUAAUGAAUUGUUGCAGGCAAAGAUGCUGCCCAAUAUGACAAAAAGGCGAUGCAAUAACUCCUCACUGCCCAUGGCCUUGGACAGUGGGAAUAUGCUGCGUA ...........(((((.......((.((((((((((((....))))(((..............)))..)))))))).))(((((((.(((......))).))))))).....)))))... ( -37.44) >DroMoj_CAF1 107262 120 + 1 CUAUGGGCGAGGCAGCCGACGGCGAGGCAUUGCCCGCAAAUAUGCUGCCCAAUAUGGCAAACAAGCGAUGCAGCAGCUGCUCGCUGUUCAGGGACUUGGACAUCGGAAAGAUGCUGCGUA ....((((((.((.(((...)))...)).))))))(((....)))((((......))))........((((((((.((..(((.(((((((....))))))).)))..)).)))))))). ( -47.50) >DroPer_CAF1 1621 120 + 1 CAAUGGGCGAGGCGGCCGACAGCGAGUUAUUGAAGGCAAAGAUGCUACCCAAUAUGGCGAAGAGGCGAUGCAGCAGCUCCUCGCUGCUCAUCGCCUUCGACAGCGGGAAGAUGCUGCGUA .....(((......)))(.(((((..........((((....)))).(((....((.(((..((((((((.((((((.....))))))))))))))))).))..)))....))))))... ( -46.60) >consensus CAAUGGGCGAGGCAGCCGACAGCGAGUUAUUGCAGGCAAAGAUGCUGCCCAAUAUGGCAAACAGGCGAUGCAGCAGCUCCUCGCUGCUCAUCGCCUUCGACAGCGGGAAGAUGCUGCGUA ........(.((((((.(((.....))).(((....)))....))))))).................((((((((.((.(((((((..............))))))).)).)))))))). (-25.53 = -25.92 + 0.39)

| Location | 330,384 – 330,504 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.28 |
| Mean single sequence MFE | -41.06 |
| Consensus MFE | -24.06 |
| Energy contribution | -24.07 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 330384 120 - 22224390 UGAGGAGCAUCUUUCCGCUAACGAAAGCCAUGAGCGGCGAGGAACUGCUGCACCGCCUGUUUGCUAUCCUGGGCAGCAUUUUCGCAUGCAAUGGAUCCUUAACGGCUGCCUCCCCCAUUG .(((((((..(((((.......)))))((((..((((((((((..((((((.(((..............)))))))))))))))).))).))))..........))).))))........ ( -38.14) >DroPse_CAF1 1637 120 - 1 UACGCAGCAUCUUCCCGCUGUCGAAGGCGAUGAGCAGCGAGGAGCUGCUGCAUCGCCUCUUCGCCAUAUUGGGCAGCAUCUUUGCCUUCAAUAACUCGCUGUCGGCCGCCUCGCCCAUUG ...(((((..........((.(((((((((((((((((.....)))))).))))))))..))).))((((((((((.....)))))..)))))....))))).(((......)))..... ( -45.90) >DroGri_CAF1 39492 120 - 1 UACGCAGCAUAUUCCCGCUGUCAAAAUCGCUGAACAGCGAACAGCUGCUGCAUCGUUUAUUUGCCAUAUUGGGCAGCAUAUUUGCUGGCAACAAUUCACCAUCGGCUGCAUCGCCGAUUG ...((((((.......(((((.....(((((....))))))))))))))))..........((((......))))(((....)))((....)).......((((((......)))))).. ( -38.71) >DroWil_CAF1 1611 120 - 1 UACGCAGCAUAUUCCCACUGUCCAAGGCCAUGGGCAGUGAGGAGUUAUUGCAUCGCCUUUUUGUCAUAUUGGGCAGCAUCUUUGCCUGCAACAAUUCAUUAUCAGCCGCCUCGCCGAUUG ...((((((.((((((((((((((......))))))))).)))))...)))...((((..(......)..)))).(((....))).)))...........(((.((......)).))).. ( -36.80) >DroMoj_CAF1 107262 120 - 1 UACGCAGCAUCUUUCCGAUGUCCAAGUCCCUGAACAGCGAGCAGCUGCUGCAUCGCUUGUUUGCCAUAUUGGGCAGCAUAUUUGCGGGCAAUGCCUCGCCGUCGGCUGCCUCGCCCAUAG ...((.(((((.....)))))..........(((((((((((((...)))).)))).))))))).....(((((.(((....)))(((((..(((........)))))))).)))))... ( -41.70) >DroPer_CAF1 1621 120 - 1 UACGCAGCAUCUUCCCGCUGUCGAAGGCGAUGAGCAGCGAGGAGCUGCUGCAUCGCCUCUUCGCCAUAUUGGGUAGCAUCUUUGCCUUCAAUAACUCGCUGUCGGCCGCCUCGCCCAUUG ...(((((.....((((.((.(((((((((((((((((.....)))))).))))))))..))).))...))))..(((....)))............))))).(((......)))..... ( -45.10) >consensus UACGCAGCAUCUUCCCGCUGUCGAAGGCCAUGAGCAGCGAGGAGCUGCUGCAUCGCCUCUUUGCCAUAUUGGGCAGCAUCUUUGCCUGCAAUAACUCGCUAUCGGCCGCCUCGCCCAUUG ...((((((..((((((((((............)))))).)))).))))))...(((.....(((......))).(((....)))..................))).............. (-24.06 = -24.07 + 0.01)

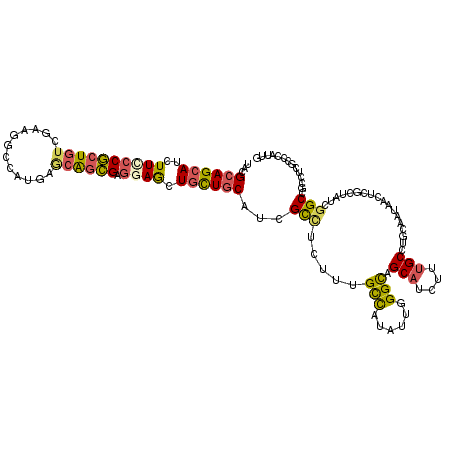
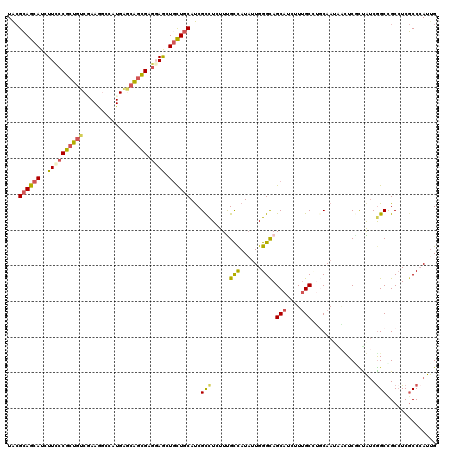
| Location | 330,424 – 330,538 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.62 |
| Mean single sequence MFE | -38.70 |
| Consensus MFE | -26.35 |
| Energy contribution | -27.02 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 330424 114 + 22224390 AAUGCUGCCCAGGAUAGCAAACAGGCGGUGCAGCAGUUCCUCGCCGCUCAUGGCUUUCGUUAGCGGAAAGAUGCUCCUCAUGGCCAGUAGAACUUGAGCGUAUCUGAAG---GAUCA- ..((((((((......((......)))).))))))(.(((((.(((((.(((.....))).)))))..((((((..((((.(..(....)..).)))).))))))).))---)).).- ( -35.00) >DroPse_CAF1 1677 114 + 1 GAUGCUGCCCAAUAUGGCGAAGAGGCGAUGCAGCAGCUCCUCGCUGCUCAUCGCCUUCGACAGCGGGAAGAUGCUGCGUAUGGCCAGCAGAACUUGAGCGUAGCUAGAA---GAUUA- ..(((((.(((.((((.(((..((((((((.((((((.....))))))))))))))))).(((((......)))))))))))).)))))...(((.(((...)))..))---)....- ( -47.70) >DroGri_CAF1 39532 113 + 1 UAUGCUGCCCAAUAUGGCAAAUAAACGAUGCAGCAGCUGUUCGCUGUUCAGCGAUUUUGACAGCGGGAAUAUGCUGCGUAUGGCUAGCAAAACUUGAGCGUAACUG----CAAAUAU- (((((((((......)))...((..((((((((((....((((((((.(((.....)))))))))))....)))))))).))..))..........))))))....----.......- ( -35.00) >DroWil_CAF1 1651 116 + 1 GAUGCUGCCCAAUAUGACAAAAAGGCGAUGCAAUAACUCCUCACUGCCCAUGGCCUUGGACAGUGGGAAUAUGCUGCGUAUGGCCAGCAGAACUUGAGCGUAGCUAAAAAG-AAAAU- ...(((((...............(((.(((((.((..(((.(((((.(((......))).))))))))...)).)))))...))).((.........))))))).......-.....- ( -31.70) >DroMoj_CAF1 107302 118 + 1 UAUGCUGCCCAAUAUGGCAAACAAGCGAUGCAGCAGCUGCUCGCUGUUCAGGGACUUGGACAUCGGAAAGAUGCUGCGUAUGGCCAGUAGAACUUGUGCAUAGCUAUCGGGAUAUCAA ..(((((.(((.....((......)).((((((((.((..(((.(((((((....))))))).)))..)).)))))))).))).)))))...((((.((...))...))))....... ( -36.50) >DroPer_CAF1 1661 114 + 1 GAUGCUACCCAAUAUGGCGAAGAGGCGAUGCAGCAGCUCCUCGCUGCUCAUCGCCUUCGACAGCGGGAAGAUGCUGCGUAUGGCCAGCAGAACUUGAGCGUAGCUAGAA---GAUUA- ...(((((......((((...(((((((((.((((((.....)))))))))))))))((.(((((......)))))))....))))((.........))))))).....---.....- ( -46.30) >consensus GAUGCUGCCCAAUAUGGCAAACAGGCGAUGCAGCAGCUCCUCGCUGCUCAUCGCCUUCGACAGCGGGAAGAUGCUGCGUAUGGCCAGCAGAACUUGAGCGUAGCUAAAA___GAUAA_ ..(((((.(((.....((......)).((((((((.((.(((((((..............))))))).)).)))))))).))).)))))............................. (-26.35 = -27.02 + 0.67)
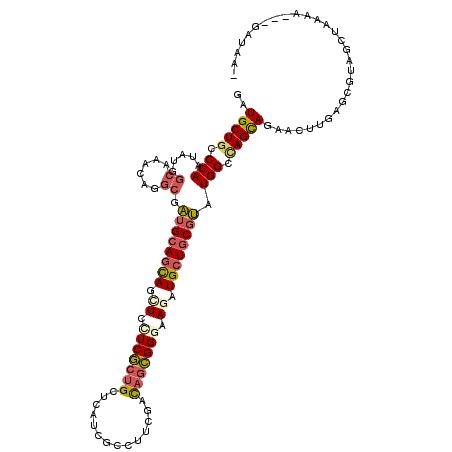
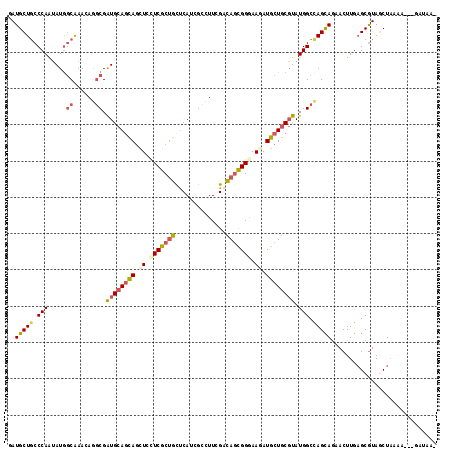
| Location | 330,424 – 330,538 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.62 |
| Mean single sequence MFE | -37.32 |
| Consensus MFE | -25.46 |
| Energy contribution | -25.97 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 330424 114 - 22224390 -UGAUC---CUUCAGAUACGCUCAAGUUCUACUGGCCAUGAGGAGCAUCUUUCCGCUAACGAAAGCCAUGAGCGGCGAGGAACUGCUGCACCGCCUGUUUGCUAUCCUGGGCAGCAUU -...((---(((..(...((((((.(((((.(.......).)))))..(((((.......)))))...)))))).))))))..((((((.(((..............))))))))).. ( -31.44) >DroPse_CAF1 1677 114 - 1 -UAAUC---UUCUAGCUACGCUCAAGUUCUGCUGGCCAUACGCAGCAUCUUCCCGCUGUCGAAGGCGAUGAGCAGCGAGGAGCUGCUGCAUCGCCUCUUCGCCAUAUUGGGCAGCAUC -.....---.....(((..((((((.......((((....((((((........)))).)).((((((((((((((.....)))))).))))))))....))))..)))))))))... ( -46.30) >DroGri_CAF1 39532 113 - 1 -AUAUUUG----CAGUUACGCUCAAGUUUUGCUAGCCAUACGCAGCAUAUUCCCGCUGUCAAAAUCGCUGAACAGCGAACAGCUGCUGCAUCGUUUAUUUGCCAUAUUGGGCAGCAUA -..(((((----.((.....)))))))..((((.(((....((((((.......(((((.....(((((....))))))))))))))))...((......)).......))))))).. ( -31.81) >DroWil_CAF1 1651 116 - 1 -AUUUU-CUUUUUAGCUACGCUCAAGUUCUGCUGGCCAUACGCAGCAUAUUCCCACUGUCCAAGGCCAUGGGCAGUGAGGAGUUAUUGCAUCGCCUUUUUGUCAUAUUGGGCAGCAUC -.....-.......((((.((.........)))))).....((.(((.((((((((((((((......))))))))).)))))...)))...((((..(......)..)))).))... ( -37.60) >DroMoj_CAF1 107302 118 - 1 UUGAUAUCCCGAUAGCUAUGCACAAGUUCUACUGGCCAUACGCAGCAUCUUUCCGAUGUCCAAGUCCCUGAACAGCGAGCAGCUGCUGCAUCGCUUGUUUGCCAUAUUGGGCAGCAUA .((....(((((((((...))...........((((........(((((.....)))))..........(((((((((((((...)))).)))).))))))))))))))))...)).. ( -30.70) >DroPer_CAF1 1661 114 - 1 -UAAUC---UUCUAGCUACGCUCAAGUUCUGCUGGCCAUACGCAGCAUCUUCCCGCUGUCGAAGGCGAUGAGCAGCGAGGAGCUGCUGCAUCGCCUCUUCGCCAUAUUGGGUAGCAUC -.....---.....(((((((.........))((((....((((((........)))).)).((((((((((((((.....)))))).))))))))....))))......)))))... ( -46.10) >consensus _UAAUC___UUCUAGCUACGCUCAAGUUCUGCUGGCCAUACGCAGCAUCUUCCCGCUGUCGAAGGCCAUGAGCAGCGAGGAGCUGCUGCAUCGCCUCUUUGCCAUAUUGGGCAGCAUC .............................(((((.(((...((((((..((((((((((............)))))).)))).))))))...((......)).....))).))))).. (-25.46 = -25.97 + 0.51)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:29 2006