
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,884,472 – 19,884,652 |
| Length | 180 |
| Max. P | 0.970505 |

| Location | 19,884,472 – 19,884,576 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.70 |
| Mean single sequence MFE | -28.88 |
| Consensus MFE | -22.23 |
| Energy contribution | -22.11 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19884472 104 + 22224390 AAACGGUUUUCCCCGCCCGCUUUUG----------------CCAGCACAAAAAAAAUGAGGCAGAAAUAAGGAAAGAAAUAACAUUUCGACAGCUGCCCCACUGCGAAAGGUGGGGCUAC ....((((.((.....((..(((((----------------((................)))))))....))...(((((...))))))).))))((((((((......))))))))... ( -28.39) >DroSec_CAF1 38183 102 + 1 AAACGGUUUUCCCCGCCCGCUUUUC----------------CCAGCACAAAAAAAAUGAGGCAGAAAUAAGGAAAGAAAUAACAUUUCGACAGCUGCCCCACUGCGA-AGGUGGGGCAA- ..........(((((((.(((....----------------..)))..........((.(((((......((((.(......).)))).....))))).))......-.)))))))...- ( -26.60) >DroSim_CAF1 28917 101 + 1 AAACGGUUUUCCCCGCCCGCUUUUC----------------CCAGCACAAAAAAAAUGAGGCAGAAAUAAGGAAAGAAAUAACAUUUCGACAGCUGCCCCACUGCGA-AGGUGG-GCAA- ....((......))(((((((((.(----------------.(((...........((.(((((......((((.(......).)))).....))))).))))).))-))))))-))..- ( -27.30) >DroEre_CAF1 17984 100 + 1 AAACGGUUUUCCCCGCCCGCUUU-----------------UCCAGCACAAA--AAAUGAGGCAGAAAUAAGGAAAGAAAUAACAUUUCGACAGCUGCCCCAUUGAAA-AGGUGGGGCUAC ..........(((((((.(((..-----------------...))).....--.((((.(((((......((((.(......).)))).....))))).))))....-.))))))).... ( -28.60) >DroYak_CAF1 14686 117 + 1 AAACGGUUUUCCCCGCCCGCUUUUCCCUUUUCUUCCUUUUUCCAGCGCAAA--AAAUGAGGCAGAAAUAAGGAAAGAAAUAACAUUUCGACAGCUGCCCCACUGAGA-AAGUGGGGCUCC ...(((......)))...((((((((..(((((.((((.............--....)))).)))))...)))))(((((...)))))...))).((((((((....-.))))))))... ( -33.53) >consensus AAACGGUUUUCCCCGCCCGCUUUUC________________CCAGCACAAAAAAAAUGAGGCAGAAAUAAGGAAAGAAAUAACAUUUCGACAGCUGCCCCACUGCGA_AGGUGGGGCUAC ....((((.((...(((.(((......................))).((.......)).))).............(((((...))))))).))))((((((((......))))))))... (-22.23 = -22.11 + -0.12)

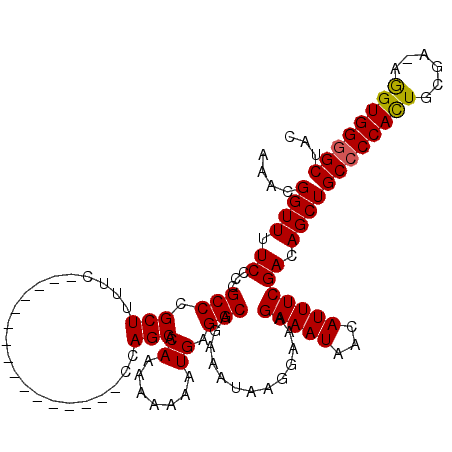

| Location | 19,884,472 – 19,884,576 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.70 |
| Mean single sequence MFE | -32.14 |
| Consensus MFE | -25.83 |
| Energy contribution | -25.55 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19884472 104 - 22224390 GUAGCCCCACCUUUCGCAGUGGGGCAGCUGUCGAAAUGUUAUUUCUUUCCUUAUUUCUGCCUCAUUUUUUUUGUGCUGG----------------CAAAAGCGGGCGGGGAAAACCGUUU ...(((((((........))))))).(((...((((((.............))))))((((.(((.......)))..))----------------))..)))((((((......)))))) ( -32.42) >DroSec_CAF1 38183 102 - 1 -UUGCCCCACCU-UCGCAGUGGGGCAGCUGUCGAAAUGUUAUUUCUUUCCUUAUUUCUGCCUCAUUUUUUUUGUGCUGG----------------GAAAAGCGGGCGGGGAAAACCGUUU -(((((((((..-.....))))))))).....(((((...)))))........(..((((((..(((((((......))----------------)))))..))))))..)......... ( -30.30) >DroSim_CAF1 28917 101 - 1 -UUGC-CCACCU-UCGCAGUGGGGCAGCUGUCGAAAUGUUAUUUCUUUCCUUAUUUCUGCCUCAUUUUUUUUGUGCUGG----------------GAAAAGCGGGCGGGGAAAACCGUUU -..((-((....-....((((((((((..((.((((.(......)))))...))..))))))))))........(((..----------------....)))))))((......)).... ( -28.60) >DroEre_CAF1 17984 100 - 1 GUAGCCCCACCU-UUUCAAUGGGGCAGCUGUCGAAAUGUUAUUUCUUUCCUUAUUUCUGCCUCAUUU--UUUGUGCUGGA-----------------AAAGCGGGCGGGGAAAACCGUUU ....((((.(((-....((((((((((..((.((((.(......)))))...))..)))))))))).--.....(((...-----------------..)))))).)))).......... ( -29.30) >DroYak_CAF1 14686 117 - 1 GGAGCCCCACUU-UCUCAGUGGGGCAGCUGUCGAAAUGUUAUUUCUUUCCUUAUUUCUGCCUCAUUU--UUUGCGCUGGAAAAAGGAAGAAAAGGGAAAAGCGGGCGGGGAAAACCGUUU ...((((((((.-....)))))))).(((...(((((...)))))(((((((.(((((.(((..(((--(((.....))))))))).)))))))))))))))((((((......)))))) ( -40.10) >consensus GUAGCCCCACCU_UCGCAGUGGGGCAGCUGUCGAAAUGUUAUUUCUUUCCUUAUUUCUGCCUCAUUUUUUUUGUGCUGG________________GAAAAGCGGGCGGGGAAAACCGUUU ....((((.((......((((((((((..((.((((.(......)))))...))..)))))))))).......((((......................)))))).)))).......... (-25.83 = -25.55 + -0.28)


| Location | 19,884,497 – 19,884,612 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.98 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -26.16 |
| Energy contribution | -26.08 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19884497 115 + 22224390 -CCAGCACAAAAAAAAUGAGGCAGAAAUAAGGAAAGAAAUAACAUUUCGACAGCUGCCCCACUGCGAAAGGUGGGGCUAC----UGGGGGUGGCCUUUCCUCUGGGUCAAUGAGCAUCAG -...............(((..((((.....((((((......(((..(..(((..((((((((......))))))))..)----)))..)))..))))))))))..)))........... ( -35.70) >DroSec_CAF1 38208 109 + 1 -CCAGCACAAAAAAAAUGAGGCAGAAAUAAGGAAAGAAAUAACAUUUCGACAGCUGCCCCACUGCGA-AGGUGGGGCAA---------GGGGGCUUUUCCUCUGGGUCAAUGAGCAUCAG -...............(((.((.............(((((...)))))(((...(((((((((....-.)))))))))(---------(((((....))))))..))).....)).))). ( -32.40) >DroSim_CAF1 28942 108 + 1 -CCAGCACAAAAAAAAUGAGGCAGAAAUAAGGAAAGAAAUAACAUUUCGACAGCUGCCCCACUGCGA-AGGUGG-GCAA---------GGGAGCUUUUCCUCUGGGUCAAUGAGCAUCAG -((.((.............(((((......((((.(......).)))).....)))))(((((....-.)))))-))..---------.)).((((..((....)).....))))..... ( -24.30) >DroEre_CAF1 18007 108 + 1 UCCAGCACAAA--AAAUGAGGCAGAAAUAAGGAAAGAAAUAACAUUUCGACAGCUGCCCCAUUGAAA-AGGUGGGGCUAC---------GGGGGUUUUCCUCUGGGUCAAUGAGCAUCAG ...........--...(((.((.............(((((...)))))(((....((((((((....-.))))))))..(---------(((((....)))))).))).....)).))). ( -31.20) >DroYak_CAF1 14726 117 + 1 UCCAGCGCAAA--AAAUGAGGCAGAAAUAAGGAAAGAAAUAACAUUUCGACAGCUGCCCCACUGAGA-AAGUGGGGCUCCAGUGUGGAGGGGGGUUUUCCUCUGGGUCAAUGAGCAUCAG ...........--...(((.((.............(((((...)))))(((....((((((((....-.)))))))).((((...(((((.....))))).))))))).....)).))). ( -34.90) >consensus _CCAGCACAAAAAAAAUGAGGCAGAAAUAAGGAAAGAAAUAACAUUUCGACAGCUGCCCCACUGCGA_AGGUGGGGCUAC________GGGGGCUUUUCCUCUGGGUCAAUGAGCAUCAG ................(((.((.............(((((...)))))(((....((((((((......))))))))............((((......))))..))).....)).))). (-26.16 = -26.08 + -0.08)


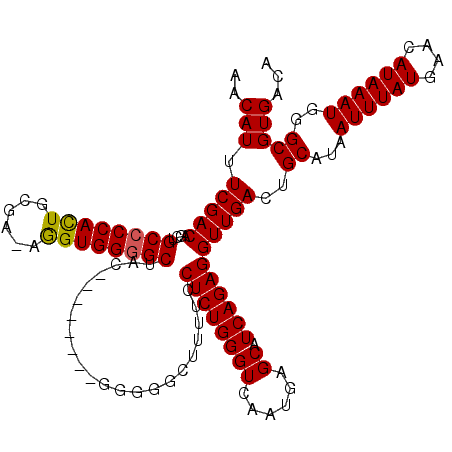
| Location | 19,884,536 – 19,884,652 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.92 |
| Mean single sequence MFE | -37.52 |
| Consensus MFE | -32.54 |
| Energy contribution | -32.42 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19884536 116 + 22224390 AACAUUUCGACAGCUGCCCCACUGCGAAAGGUGGGGCUAC----UGGGGGUGGCCUUUCCUCUGGGUCAAUGAGCAUCAGAGGUUGACUGCAUAAUUUAUGAACAUAAAUGGGCGUGACA ..(((..(..(((..((((((((......))))))))..)----)))..)))((((..(((((((((......)).)))))))...........((((((....))))))))))...... ( -41.90) >DroSec_CAF1 38247 110 + 1 AACAUUUCGACAGCUGCCCCACUGCGA-AGGUGGGGCAA---------GGGGGCUUUUCCUCUGGGUCAAUGAGCAUCAGAGGUUGACUGCAUAAUUUAUGAACAUAAAUGGGCGUGACA ..(((.(((((((((((((((((....-.))))))))..---------...))))....((((((((......)).)))))))))))..((...((((((....))))))..)))))... ( -35.50) >DroSim_CAF1 28981 109 + 1 AACAUUUCGACAGCUGCCCCACUGCGA-AGGUGG-GCAA---------GGGAGCUUUUCCUCUGGGUCAAUGAGCAUCAGAGGUUGACUGCAUAAUUUAUGAACAUAAAUGGGCGUGACA ..(((.(((((((((.(((..(..(..-..)..)-....---------)))))))....((((((((......)).)))))))))))..((...((((((....))))))..)))))... ( -31.30) >DroEre_CAF1 18045 110 + 1 AACAUUUCGACAGCUGCCCCAUUGAAA-AGGUGGGGCUAC---------GGGGGUUUUCCUCUGGGUCAAUGAGCAUCAGAGGUUGACUGCAUAAUUUAUGAACAUAAAUGGGCGUGACA ......(((...(((((((((((....-.))))))))...---------.(.((((..(((((((((......)).)))))))..)))).)...((((((....)))))).))).))).. ( -36.10) >DroYak_CAF1 14764 119 + 1 AACAUUUCGACAGCUGCCCCACUGAGA-AAGUGGGGCUCCAGUGUGGAGGGGGGUUUUCCUCUGGGUCAAUGAGCAUCAGAGGUUGACUGCAUAAUUUAUGAACAUAAAUGGGCGUGACA ....((((.(((.((((((((((....-.))))))))...))))).))))(.((((..(((((((((......)).)))))))..)))).).....(((((..((....))..))))).. ( -42.80) >consensus AACAUUUCGACAGCUGCCCCACUGCGA_AGGUGGGGCUAC________GGGGGCUUUUCCUCUGGGUCAAUGAGCAUCAGAGGUUGACUGCAUAAUUUAUGAACAUAAAUGGGCGUGACA ..(((.(((((....((((((((......))))))))......................((((((((......)).)))))))))))..((...((((((....))))))..)))))... (-32.54 = -32.42 + -0.12)


| Location | 19,884,536 – 19,884,652 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.92 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -27.10 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19884536 116 - 22224390 UGUCACGCCCAUUUAUGUUCAUAAAUUAUGCAGUCAACCUCUGAUGCUCAUUGACCCAGAGGAAAGGCCACCCCCA----GUAGCCCCACCUUUCGCAGUGGGGCAGCUGUCGAAAUGUU .(((..((..((((((....))))))...))......((((((...(.....)...))))))...)))...(..((----((.(((((((........))))))).))))..)....... ( -34.20) >DroSec_CAF1 38247 110 - 1 UGUCACGCCCAUUUAUGUUCAUAAAUUAUGCAGUCAACCUCUGAUGCUCAUUGACCCAGAGGAAAAGCCCCC---------UUGCCCCACCU-UCGCAGUGGGGCAGCUGUCGAAAUGUU ..((......((((((....))))))...(((((...((((((...(.....)...))))))..........---------.((((((((..-.....))))))))))))).))...... ( -29.80) >DroSim_CAF1 28981 109 - 1 UGUCACGCCCAUUUAUGUUCAUAAAUUAUGCAGUCAACCUCUGAUGCUCAUUGACCCAGAGGAAAAGCUCCC---------UUGC-CCACCU-UCGCAGUGGGGCAGCUGUCGAAAUGUU ..((......((((((....))))))...(((((...((((((...(.....)...))))))....(((((.---------((((-......-..)))).))))).))))).))...... ( -27.00) >DroEre_CAF1 18045 110 - 1 UGUCACGCCCAUUUAUGUUCAUAAAUUAUGCAGUCAACCUCUGAUGCUCAUUGACCCAGAGGAAAACCCCC---------GUAGCCCCACCU-UUUCAAUGGGGCAGCUGUCGAAAUGUU ..((......((((((....))))))...(((((...((((((...(.....)...)))))).........---------...((((((...-......)))))).))))).))...... ( -25.50) >DroYak_CAF1 14764 119 - 1 UGUCACGCCCAUUUAUGUUCAUAAAUUAUGCAGUCAACCUCUGAUGCUCAUUGACCCAGAGGAAAACCCCCCUCCACACUGGAGCCCCACUU-UCUCAGUGGGGCAGCUGUCGAAAUGUU ..((......((((((....))))))...(((((...((((((...(.....)...))))))..........(((.....)))((((((((.-....)))))))).))))).))...... ( -32.50) >consensus UGUCACGCCCAUUUAUGUUCAUAAAUUAUGCAGUCAACCUCUGAUGCUCAUUGACCCAGAGGAAAAGCCCCC________GUAGCCCCACCU_UCGCAGUGGGGCAGCUGUCGAAAUGUU ..((......((((((....))))))...(((((...((((((...(.....)...)))))).....................(((((((........))))))).))))).))...... (-27.10 = -27.50 + 0.40)

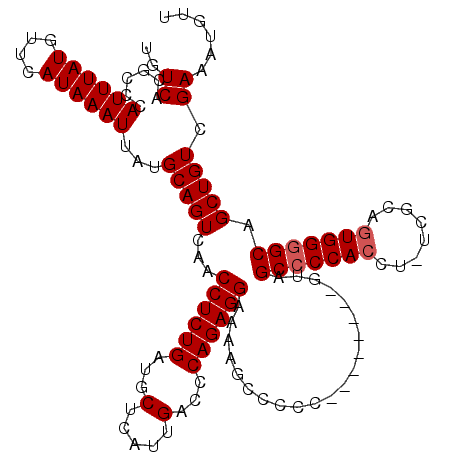
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:46 2006