
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,884,020 – 19,884,214 |
| Length | 194 |
| Max. P | 0.987926 |

| Location | 19,884,020 – 19,884,135 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.41 |
| Mean single sequence MFE | -34.24 |
| Consensus MFE | -30.52 |
| Energy contribution | -30.52 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19884020 115 + 22224390 CGAGUCCAUUGUACGUGCUUCGUUGGCGGGACUGCGGGAAUCAAACCAGAAACCAGAAAACCAAACUUUCAAUGGACUG---GCUGGCAGCAAGUCAAGCUGCCCAAAAACGGAAAAC-- (.(((((((((...(((((.....)))((.......((..((......))..))......))..))...))))))))).---)..((((((.......))))))..............-- ( -31.42) >DroSec_CAF1 37742 113 + 1 AGAGUCCAUUGUACGUGCUUCGUUGGCGGAGCUGCGGGAACC--ACCAGAAACCAGAAAACAAAACUUUCAAUGGACUG---GCUGGCAGCAAGUCAAGCUGCCCGAAAACAGAAAAC-- ..(((((((((..(((((((((....)))))).)))((....--.))......................))))))))).---.(.((((((.......)))))).)............-- ( -35.80) >DroSim_CAF1 28477 113 + 1 CGAGUCCAUUGUACGUGCUUCGUUGGCGGAGCUGCGGGAACC--ACCAGAAACCAGAAAACAAAACUUUCAAUGGACUG---GCUGGCAGCAAGUCAAGCUGCCCGAAAACAGAAAAC-- (.(((((((((..(((((((((....)))))).)))((....--.))......................))))))))).---)(.((((((.......)))))).)............-- ( -36.60) >DroEre_CAF1 17547 103 + 1 CAAGUCCAUUGUACGUGCUUCGUUGGCGGGACGGAGG------------AACCCAGAAAACAAAACUUUCAAUGGACUG---GCUGGCAGCAAGUCAAGCUGCCCAAAAUCAGAAAAC-- ..(((((((((...(((((.....)))(((.......------------..)))..........))...))))))))).---...((((((.......))))))..............-- ( -29.90) >DroYak_CAF1 14235 108 + 1 CGAGUCCAUUGUACGUGCUCCGUUGGCGGGGCUGGGG------------AAUCCAGAAAACAAAACUUUCAAUGGACUGACGGCAGGCAGCAAGUCAAGCUGCCCGAAAUCAGAAAACAA ..(((((((((...((((((((....))))))(((..------------...))).........))...)))))))))((...(.((((((.......)))))).)...))......... ( -37.50) >consensus CGAGUCCAUUGUACGUGCUUCGUUGGCGGGGCUGCGGGAA_C__ACCAGAAACCAGAAAACAAAACUUUCAAUGGACUG___GCUGGCAGCAAGUCAAGCUGCCCGAAAACAGAAAAC__ ..(((((((((...((((((((....))))))...((...............))..........))...))))))))).......((((((.......))))))................ (-30.52 = -30.52 + -0.00)



| Location | 19,884,020 – 19,884,135 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.41 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -29.22 |
| Energy contribution | -29.26 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19884020 115 - 22224390 --GUUUUCCGUUUUUGGGCAGCUUGACUUGCUGCCAGC---CAGUCCAUUGAAAGUUUGGUUUUCUGGUUUCUGGUUUGAUUCCCGCAGUCCCGCCAACGAAGCACGUACAAUGGACUCG --..............((((((.......))))))...---.(((((((((.......((....)).(((((((((..((((.....))))..))))..))))).....))))))))).. ( -33.10) >DroSec_CAF1 37742 113 - 1 --GUUUUCUGUUUUCGGGCAGCUUGACUUGCUGCCAGC---CAGUCCAUUGAAAGUUUUGUUUUCUGGUUUCUGGU--GGUUCCCGCAGCUCCGCCAACGAAGCACGUACAAUGGACUCU --..............((((((.......))))))...---.((((((((((..((((((((...(((...(((.(--((...))))))..)))..))))))))...).))))))))).. ( -36.00) >DroSim_CAF1 28477 113 - 1 --GUUUUCUGUUUUCGGGCAGCUUGACUUGCUGCCAGC---CAGUCCAUUGAAAGUUUUGUUUUCUGGUUUCUGGU--GGUUCCCGCAGCUCCGCCAACGAAGCACGUACAAUGGACUCG --..............((((((.......))))))...---.((((((((((..((((((((...(((...(((.(--((...))))))..)))..))))))))...).))))))))).. ( -36.00) >DroEre_CAF1 17547 103 - 1 --GUUUUCUGAUUUUGGGCAGCUUGACUUGCUGCCAGC---CAGUCCAUUGAAAGUUUUGUUUUCUGGGUU------------CCUCCGUCCCGCCAACGAAGCACGUACAAUGGACUUG --..............((((((.......))))))...---.((((((((((..((((((((...((((..------------.......))))..))))))))...).))))))))).. ( -34.10) >DroYak_CAF1 14235 108 - 1 UUGUUUUCUGAUUUCGGGCAGCUUGACUUGCUGCCUGCCGUCAGUCCAUUGAAAGUUUUGUUUUCUGGAUU------------CCCCAGCCCCGCCAACGGAGCACGUACAAUGGACUCG ........((((..((((((((.......))))))))..))))((((((((..............(((...------------..)))((.(((....))).)).....))))))))... ( -38.70) >consensus __GUUUUCUGUUUUCGGGCAGCUUGACUUGCUGCCAGC___CAGUCCAUUGAAAGUUUUGUUUUCUGGUUUCUGGU__G_UUCCCGCAGCCCCGCCAACGAAGCACGUACAAUGGACUCG ................((((((.......)))))).......((((((((((..((((((((...(((.......................)))..))))))))...).))))))))).. (-29.22 = -29.26 + 0.04)

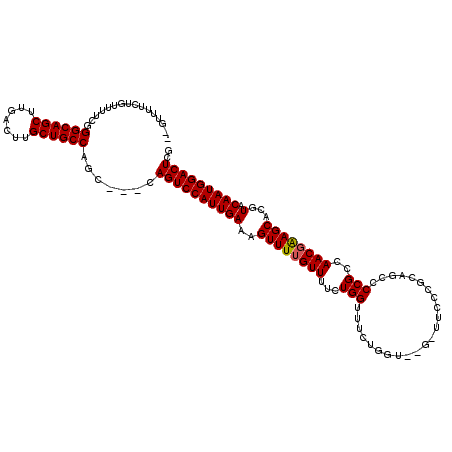
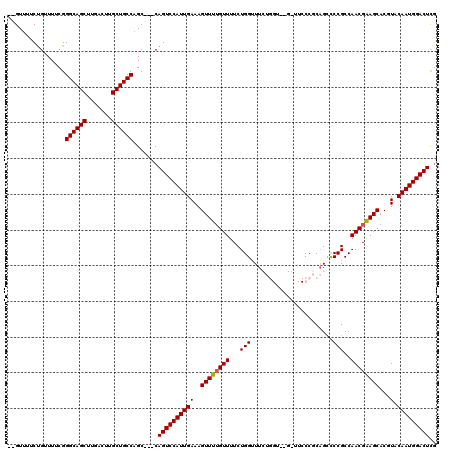
| Location | 19,884,060 – 19,884,174 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.81 |
| Mean single sequence MFE | -25.96 |
| Consensus MFE | -21.52 |
| Energy contribution | -21.72 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19884060 114 + 22224390 UCAAACCAGAAACCAGAAAACCAAACUUUCAAUGGACUG---GCUGGCAGCAAGUCAAGCUGCCCAAAAACGGAAAAC---AAAAAAUCCCAAUCGACUGGGGAUGGUAUCCAUCUUUUU .....((.....((((....(((.........))).)))---)..((((((.......)))))).......)).....---.......((((......))))(((((...)))))..... ( -27.00) >DroSec_CAF1 37782 112 + 1 CC--ACCAGAAACCAGAAAACAAAACUUUCAAUGGACUG---GCUGGCAGCAAGUCAAGCUGCCCGAAAACAGAAAAC---AAAAAAUUCCAAUCGACUGGGGAUGGUAUCCAUAUUUUU ((--.((((...(((((((.......))))..))).(((---.(.((((((.......)))))).)....))).....---................))))))((((...))))...... ( -26.50) >DroSim_CAF1 28517 112 + 1 CC--ACCAGAAACCAGAAAACAAAACUUUCAAUGGACUG---GCUGGCAGCAAGUCAAGCUGCCCGAAAACAGAAAAC---AAAAAGUCCCAAUCGACUGGGGAUGGUAUCCAUCUUUUU ..--.((((...(((((((.......))))..))).(((---.(.((((((.......)))))).)....))).....---................))))((((((...)))))).... ( -28.30) >DroEre_CAF1 17584 105 + 1 ---------AACCCAGAAAACAAAACUUUCAAUGGACUG---GCUGGCAGCAAGUCAAGCUGCCCAAAAUCAGAAAAC---AAAAAAUACGAAUUGACUGGGGAUGGUAUCCAACCUUUU ---------..(((((.........((......)).(((---(..((((((.......)))))).....)))).....---................)))))...(((.....))).... ( -23.10) >DroYak_CAF1 14272 111 + 1 ---------AAUCCAGAAAACAAAACUUUCAAUGGACUGACGGCAGGCAGCAAGUCAAGCUGCCCGAAAUCAGAAAACAACAAAAAAUACCAAUUGACUGGGGAUUGUAUCUAUCCUUUU ---------..((((((((.......))))..))))((((...(.((((((.......)))))).)...))))................(((......)))((((.......)))).... ( -24.90) >consensus _C__ACCAGAAACCAGAAAACAAAACUUUCAAUGGACUG___GCUGGCAGCAAGUCAAGCUGCCCGAAAACAGAAAAC___AAAAAAUACCAAUCGACUGGGGAUGGUAUCCAUCUUUUU ............((((.........((......)).(((......((((((.......))))))......)))........................))))((((((...)))))).... (-21.52 = -21.72 + 0.20)

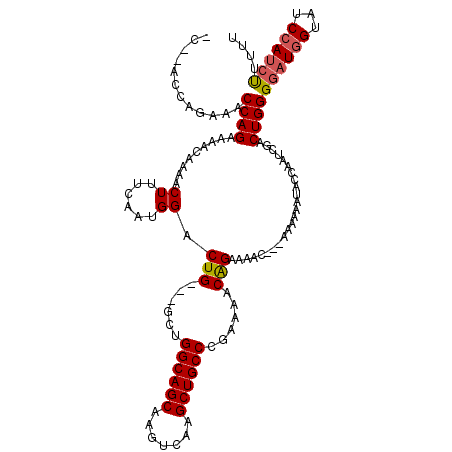
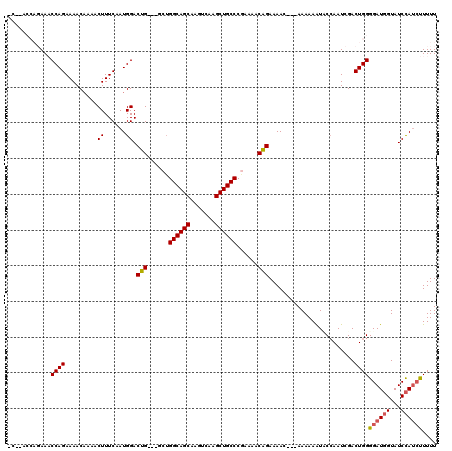
| Location | 19,884,060 – 19,884,174 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.81 |
| Mean single sequence MFE | -31.34 |
| Consensus MFE | -22.16 |
| Energy contribution | -22.56 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19884060 114 - 22224390 AAAAAGAUGGAUACCAUCCCCAGUCGAUUGGGAUUUUUU---GUUUUCCGUUUUUGGGCAGCUUGACUUGCUGCCAGC---CAGUCCAUUGAAAGUUUGGUUUUCUGGUUUCUGGUUUGA ..(((((((((...((..(((((....)))))......)---)...))))))))).((((((.......))))))(((---(((.(((..(((((.....))))))))...))))))... ( -35.50) >DroSec_CAF1 37782 112 - 1 AAAAAUAUGGAUACCAUCCCCAGUCGAUUGGAAUUUUUU---GUUUUCUGUUUUCGGGCAGCUUGACUUGCUGCCAGC---CAGUCCAUUGAAAGUUUUGUUUUCUGGUUUCUGGU--GG ...........(((((...((((..((.(..(((((((.---.....(((......((((((.......))))))...---)))......)))))))..)))..))))....))))--). ( -28.50) >DroSim_CAF1 28517 112 - 1 AAAAAGAUGGAUACCAUCCCCAGUCGAUUGGGACUUUUU---GUUUUCUGUUUUCGGGCAGCUUGACUUGCUGCCAGC---CAGUCCAUUGAAAGUUUUGUUUUCUGGUUUCUGGU--GG .....(((((...)))))(((((....))))).......---..............((((((.......)))))).((---(((.(((..(((((.....))))))))...)))))--.. ( -32.60) >DroEre_CAF1 17584 105 - 1 AAAAGGUUGGAUACCAUCCCCAGUCAAUUCGUAUUUUUU---GUUUUCUGAUUUUGGGCAGCUUGACUUGCUGCCAGC---CAGUCCAUUGAAAGUUUUGUUUUCUGGGUU--------- ....(((.....)))...(((((..(((....((((((.---.....(((......((((((.......))))))...---)))......))))))...)))..)))))..--------- ( -27.70) >DroYak_CAF1 14272 111 - 1 AAAAGGAUAGAUACAAUCCCCAGUCAAUUGGUAUUUUUUGUUGUUUUCUGAUUUCGGGCAGCUUGACUUGCUGCCUGCCGUCAGUCCAUUGAAAGUUUUGUUUUCUGGAUU--------- ....((((.......))))((((..(((....((((((...((....(((((..((((((((.......))))))))..)))))..))..))))))...)))..))))...--------- ( -32.40) >consensus AAAAAGAUGGAUACCAUCCCCAGUCGAUUGGGAUUUUUU___GUUUUCUGUUUUCGGGCAGCUUGACUUGCUGCCAGC___CAGUCCAUUGAAAGUUUUGUUUUCUGGUUUCUGGU__G_ .....(((((...))))).((((..(((..(.((((((.........(((......((((((.......))))))......)))......)))))))..)))..))))............ (-22.16 = -22.56 + 0.40)


| Location | 19,884,099 – 19,884,214 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.63 |
| Mean single sequence MFE | -24.88 |
| Consensus MFE | -18.02 |
| Energy contribution | -18.54 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19884099 115 + 22224390 --GCUGGCAGCAAGUCAAGCUGCCCAAAAACGGAAAAC---AAAAAAUCCCAAUCGACUGGGGAUGGUAUCCAUCUUUUUAAUCAAAGCCAACUCCCAAUGCAAAAUAUUGCUCUACCCA --...((((((.......)))))).......(((....---.......((((......))))(((((...)))))..................)))....((((....))))........ ( -25.20) >DroSec_CAF1 37819 115 + 1 --GCUGGCAGCAAGUCAAGCUGCCCGAAAACAGAAAAC---AAAAAAUUCCAAUCGACUGGGGAUGGUAUCCAUAUUUUUAAUCAAAGCCAACUCGCAAUGCAAAAUAUUGCUCCACCCA --.(.((((((.......)))))).)............---..................(((..((((...................))))....((((((.....))))))....))). ( -22.61) >DroSim_CAF1 28554 115 + 1 --GCUGGCAGCAAGUCAAGCUGCCCGAAAACAGAAAAC---AAAAAGUCCCAAUCGACUGGGGAUGGUAUCCAUCUUUUUAAUCAAAGCCAACUCGCAAUGCAAAAUAUUGCUCCACCCA --.(.((((((.......)))))).)............---....((((......))))((((((((...)))))....................((((((.....))))))....))). ( -27.90) >DroEre_CAF1 17614 115 + 1 --GCUGGCAGCAAGUCAAGCUGCCCAAAAUCAGAAAAC---AAAAAAUACGAAUUGACUGGGGAUGGUAUCCAACCUUUUAAUCAAAGCCAACUCCCAAUGCAAAAUAUGGUUCCAGCCA --((.((((((.......))))))..............---.................(((((.((((...................)))).).))))..))......((((....)))) ( -23.81) >DroYak_CAF1 14303 115 + 1 CGGCAGGCAGCAAGUCAAGCUGCCCGAAAUCAGAAAACAACAAAAAAUACCAAUUGACUGGGGAUUGUAUCUAUCCUUUUAAUCAAAGCCAACUCCCAAUGCAAAAUAUUGUGCU----- .(((.((((((.......)))))).............................((((..((((((.......))))))....)))).)))..........(((........))).----- ( -24.90) >consensus __GCUGGCAGCAAGUCAAGCUGCCCGAAAACAGAAAAC___AAAAAAUACCAAUCGACUGGGGAUGGUAUCCAUCUUUUUAAUCAAAGCCAACUCCCAAUGCAAAAUAUUGCUCCACCCA ..(((((((((.......))))))...............................((..((((((((...))))))))....))..)))...........((((....))))........ (-18.02 = -18.54 + 0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:42 2006