
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,883,723 – 19,883,984 |
| Length | 261 |
| Max. P | 0.999864 |

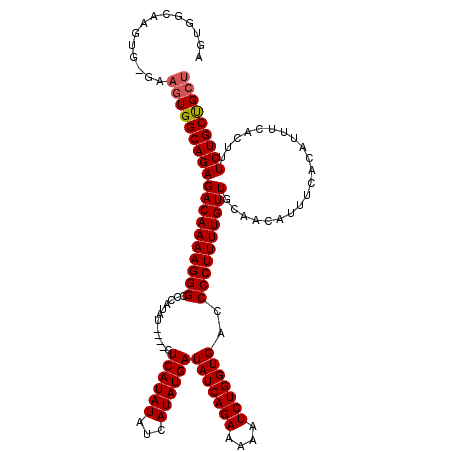
| Location | 19,883,723 – 19,883,838 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.56 |
| Mean single sequence MFE | -37.22 |
| Consensus MFE | -29.75 |
| Energy contribution | -29.99 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.30 |
| Structure conservation index | 0.80 |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.998351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19883723 115 + 22224390 AGUGGCAAGUG-GAAGUGGCAGAGACAAAAGGGCCCAUAU----CUCAUAUAUCAUAUGAUAUCAGAAAAAUCUGGUGACCCCUUUUGUUGCAACAUUUCACAUUUCACUUUCUGCUGCU ((..(((((((-(((((((....((((((((((.......----.((((((...))))))(((((((....)))))))..))))))))))........)))).)))))))...)))..)) ( -37.40) >DroSec_CAF1 37466 115 + 1 AGUGGCAAGUG-GAAGUGGCAGAGACAAAAGGGCCCAUAU----CUCAUAUAUCAUAUGAUAUCAGAAAUAUCUGGUGACCCCUUUUGUUGCAACAUUUCACAUUUCACUUUCUGCUGCU ((..(((((((-(((((((....((((((((((.......----.((((((...))))))(((((((....)))))))..))))))))))........)))).)))))))...)))..)) ( -37.40) >DroSim_CAF1 28211 115 + 1 AGUGGCAAGUG-GAAGUGGCAGAGACAAAAGGGCCCAUAU----CUCAUAUAUCAUAUGAUAUCAGAAAAAUCUGGUGACCCCUUUUGUUGCAACAUUUCACAUUUCACUUUCUGCUGCU ((..(((((((-(((((((....((((((((((.......----.((((((...))))))(((((((....)))))))..))))))))))........)))).)))))))...)))..)) ( -37.40) >DroEre_CAF1 17283 120 + 1 AGUGGGAAGUGGGAAGUGGCAGAGACAAAAGGGCCCAUAGAACACUCAUAUAUCAUAUGAUAUCAGAAAAAUCUGGUGACCCCUUUUGUUGCAACAUUUCACAUUUCACUUUCUGCUGCU ((..(((((((..(.((((....((((((((((............((((((...))))))(((((((....)))))))..))))))))))........)))).)..)))))))..))... ( -40.90) >DroYak_CAF1 13956 102 + 1 AG--------------UGGCAGAGACAAAAGGGCCCAUAU----CUCAUAUAUCAUAUGAUAUCAGAAAAAUCUGGUGACCCCUUUUGUUGCAACAUUUCACAUUUCACUUUCUGCCGCU ((--------------(((((((((((((((((.......----.((((((...))))))(((((((....)))))))..)))))))))).....................))))))))) ( -32.99) >consensus AGUGGCAAGUG_GAAGUGGCAGAGACAAAAGGGCCCAUAU____CUCAUAUAUCAUAUGAUAUCAGAAAAAUCUGGUGACCCCUUUUGUUGCAACAUUUCACAUUUCACUUUCUGCUGCU ..............(((((((((((((((((((............((((((...))))))(((((((....)))))))..)))))))))).....................))))))))) (-29.75 = -29.99 + 0.24)

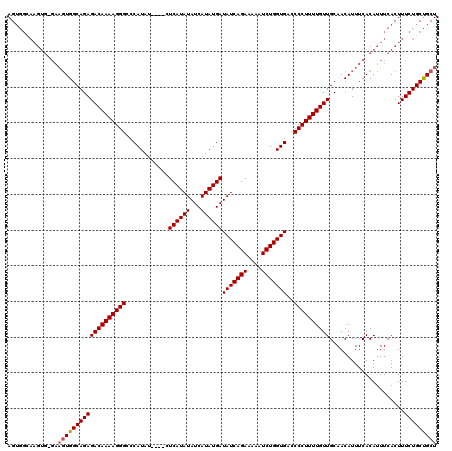
| Location | 19,883,723 – 19,883,838 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.56 |
| Mean single sequence MFE | -32.36 |
| Consensus MFE | -30.16 |
| Energy contribution | -30.16 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.30 |
| SVM RNA-class probability | 0.999864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19883723 115 - 22224390 AGCAGCAGAAAGUGAAAUGUGAAAUGUUGCAACAAAAGGGGUCACCAGAUUUUUCUGAUAUCAUAUGAUAUAUGAG----AUAUGGGCCCUUUUGUCUCUGCCACUUC-CACUUGCCACU ..........((((....(((.((.((.((((((((((((.(((.((((....))))(((((....))))).....----...))).)))))))))...))).)))).-)))....)))) ( -31.60) >DroSec_CAF1 37466 115 - 1 AGCAGCAGAAAGUGAAAUGUGAAAUGUUGCAACAAAAGGGGUCACCAGAUAUUUCUGAUAUCAUAUGAUAUAUGAG----AUAUGGGCCCUUUUGUCUCUGCCACUUC-CACUUGCCACU ..........((((....(((.((.((.((((((((((((.(((.....((((((..(((((....)))))..)))----)))))).)))))))))...))).)))).-)))....)))) ( -31.60) >DroSim_CAF1 28211 115 - 1 AGCAGCAGAAAGUGAAAUGUGAAAUGUUGCAACAAAAGGGGUCACCAGAUUUUUCUGAUAUCAUAUGAUAUAUGAG----AUAUGGGCCCUUUUGUCUCUGCCACUUC-CACUUGCCACU ..........((((....(((.((.((.((((((((((((.(((.((((....))))(((((....))))).....----...))).)))))))))...))).)))).-)))....)))) ( -31.60) >DroEre_CAF1 17283 120 - 1 AGCAGCAGAAAGUGAAAUGUGAAAUGUUGCAACAAAAGGGGUCACCAGAUUUUUCUGAUAUCAUAUGAUAUAUGAGUGUUCUAUGGGCCCUUUUGUCUCUGCCACUUCCCACUUCCCACU ..........((((....(((....((.((((((((((((.(((..(((...(((..(((((....)))))..)))...))).))).)))))))))...))).))....)))....)))) ( -32.50) >DroYak_CAF1 13956 102 - 1 AGCGGCAGAAAGUGAAAUGUGAAAUGUUGCAACAAAAGGGGUCACCAGAUUUUUCUGAUAUCAUAUGAUAUAUGAG----AUAUGGGCCCUUUUGUCUCUGCCA--------------CU ...((((((........((..(....)..))(((((((((.(((.((((....))))(((((....))))).....----...))).))))))))).)))))).--------------.. ( -34.50) >consensus AGCAGCAGAAAGUGAAAUGUGAAAUGUUGCAACAAAAGGGGUCACCAGAUUUUUCUGAUAUCAUAUGAUAUAUGAG____AUAUGGGCCCUUUUGUCUCUGCCACUUC_CACUUGCCACU ....(((((........((..(....)..))(((((((((.(((.((((....))))(((((....)))))............))).))))))))).))))).................. (-30.16 = -30.16 + 0.00)

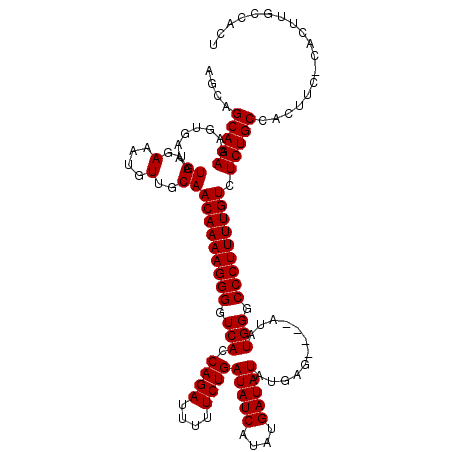
| Location | 19,883,762 – 19,883,869 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.91 |
| Mean single sequence MFE | -22.64 |
| Consensus MFE | -16.76 |
| Energy contribution | -16.76 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19883762 107 + 22224390 ----CUCAUAUAUCAUAUGAUAUCAGAAAAAUCUGGUGACCCCUUUUGUUGCAACAUUUCACAUUUCACUUUCUGCUGCUGUUUGCCGUUU---------GAAAACAAAUGCGAAAUGUU ----.((((((...))))))(((((((....)))))))..............((((((((.(((((...((((.((.((.....)).))..---------))))..))))).)))))))) ( -23.90) >DroSec_CAF1 37505 107 + 1 ----CUCAUAUAUCAUAUGAUAUCAGAAAUAUCUGGUGACCCCUUUUGUUGCAACAUUUCACAUUUCACUUUCUGCUGCUGUUUGCCGUUU---------GAAAACAAAUGCGAAAUGUU ----.((((((...))))))(((((((....)))))))..............((((((((.(((((...((((.((.((.....)).))..---------))))..))))).)))))))) ( -23.90) >DroSim_CAF1 28250 107 + 1 ----CUCAUAUAUCAUAUGAUAUCAGAAAAAUCUGGUGACCCCUUUUGUUGCAACAUUUCACAUUUCACUUUCUGCUGCUGUUUGCCGUUU---------GAAAACAAAUGCGAAAUGUU ----.((((((...))))))(((((((....)))))))..............((((((((.(((((...((((.((.((.....)).))..---------))))..))))).)))))))) ( -23.90) >DroEre_CAF1 17323 107 + 1 AACACUCAUAUAUCAUAUGAUAUCAGAAAAAUCUGGUGACCCCUUUUGUUGCAACAUUUCACAUUUCACUUUCUGCUGCUGCUUGCCGAA-------------AACAAAUGCGAAAUGUU ((((.((((((...))))))(((((((....)))))))........))))..((((((((.(((((...((((.((........)).)))-------------)..))))).)))))))) ( -24.40) >DroYak_CAF1 13982 116 + 1 ----CUCAUAUAUCAUAUGAUAUCAGAAAAAUCUGGUGACCCCUUUUGUUGCAACAUUUCACAUUUCACUUUCUGCCGCUGUUUGCCAAAAAAAAAAAAAAAAAACAAAUGCGAAAUGUU ----.((((((...))))))(((((((....)))))))..............((((((((.(((((...............(((....)))...............))))).)))))))) ( -17.11) >consensus ____CUCAUAUAUCAUAUGAUAUCAGAAAAAUCUGGUGACCCCUUUUGUUGCAACAUUUCACAUUUCACUUUCUGCUGCUGUUUGCCGUUU_________GAAAACAAAUGCGAAAUGUU .....((((((...))))))(((((((....)))))))..............((((((((.(((((..(.....)..((.....))....................))))).)))))))) (-16.76 = -16.76 + -0.00)


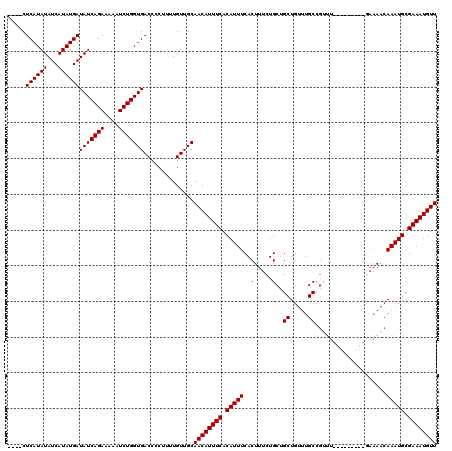
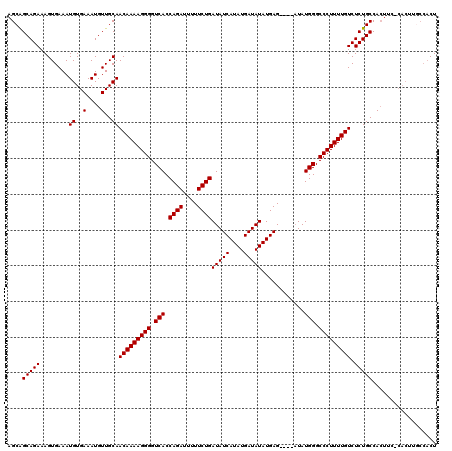
| Location | 19,883,762 – 19,883,869 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.91 |
| Mean single sequence MFE | -26.49 |
| Consensus MFE | -22.42 |
| Energy contribution | -22.42 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.991030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19883762 107 - 22224390 AACAUUUCGCAUUUGUUUUC---------AAACGGCAAACAGCAGCAGAAAGUGAAAUGUGAAAUGUUGCAACAAAAGGGGUCACCAGAUUUUUCUGAUAUCAUAUGAUAUAUGAG---- (((((((((((((..(((((---------.....((.....))....)))))..).)))))))))))).............(((.((((....))))(((((....))))).))).---- ( -26.60) >DroSec_CAF1 37505 107 - 1 AACAUUUCGCAUUUGUUUUC---------AAACGGCAAACAGCAGCAGAAAGUGAAAUGUGAAAUGUUGCAACAAAAGGGGUCACCAGAUAUUUCUGAUAUCAUAUGAUAUAUGAG---- (((((((((((((..(((((---------.....((.....))....)))))..).)))))))))))).............(((.((((....))))(((((....))))).))).---- ( -26.60) >DroSim_CAF1 28250 107 - 1 AACAUUUCGCAUUUGUUUUC---------AAACGGCAAACAGCAGCAGAAAGUGAAAUGUGAAAUGUUGCAACAAAAGGGGUCACCAGAUUUUUCUGAUAUCAUAUGAUAUAUGAG---- (((((((((((((..(((((---------.....((.....))....)))))..).)))))))))))).............(((.((((....))))(((((....))))).))).---- ( -26.60) >DroEre_CAF1 17323 107 - 1 AACAUUUCGCAUUUGUU-------------UUCGGCAAGCAGCAGCAGAAAGUGAAAUGUGAAAUGUUGCAACAAAAGGGGUCACCAGAUUUUUCUGAUAUCAUAUGAUAUAUGAGUGUU (((((((((((((..((-------------(((.((........)).)))))..).))))))))))))..............(((((((....))))(((((....)))))....))).. ( -28.70) >DroYak_CAF1 13982 116 - 1 AACAUUUCGCAUUUGUUUUUUUUUUUUUUUUUUGGCAAACAGCGGCAGAAAGUGAAAUGUGAAAUGUUGCAACAAAAGGGGUCACCAGAUUUUUCUGAUAUCAUAUGAUAUAUGAG---- ((((((((((((((.............(((((((.(.......).)))))))..)))))))))))))).............(((.((((....))))(((((....))))).))).---- ( -23.96) >consensus AACAUUUCGCAUUUGUUUUC_________AAACGGCAAACAGCAGCAGAAAGUGAAAUGUGAAAUGUUGCAACAAAAGGGGUCACCAGAUUUUUCUGAUAUCAUAUGAUAUAUGAG____ ((((((((((((((....................((.....)).((.....)).)))))))))))))).............(((.((((....))))(((((....))))).)))..... (-22.42 = -22.42 + -0.00)



| Location | 19,883,798 – 19,883,909 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -31.27 |
| Consensus MFE | -26.60 |
| Energy contribution | -26.80 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.56 |
| SVM RNA-class probability | 0.999391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19883798 111 - 22224390 CCCUAAGAACGCACACUAUUUGCUUAUGCAGAUUUCGACCAACAUUUCGCAUUUGUUUUC---------AAACGGCAAACAGCAGCAGAAAGUGAAAUGUGAAAUGUUGCAACAAAAGGG ((((.............((((((....))))))......((((((((((((((..(((((---------.....((.....))....)))))..).))))))))))))).......)))) ( -31.60) >DroSec_CAF1 37541 111 - 1 CCCCAAGAACGCACACUAUUUGCUUAUGCAGAUUUCGACCAACAUUUCGCAUUUGUUUUC---------AAACGGCAAACAGCAGCAGAAAGUGAAAUGUGAAAUGUUGCAACAAAAGGG .(((.............((((((....))))))......((((((((((((((..(((((---------.....((.....))....)))))..).)))))))))))))........))) ( -31.20) >DroSim_CAF1 28286 111 - 1 CCCUAAGAACGCACACUAUUUGCUUAUGCAGAUUUCGACCAACAUUUCGCAUUUGUUUUC---------AAACGGCAAACAGCAGCAGAAAGUGAAAUGUGAAAUGUUGCAACAAAAGGG ((((.............((((((....))))))......((((((((((((((..(((((---------.....((.....))....)))))..).))))))))))))).......)))) ( -31.60) >DroEre_CAF1 17363 107 - 1 CCCUAAGAACGCACACUAUUUGCUUAUGCAGAUUUCGACCAACAUUUCGCAUUUGUU-------------UUCGGCAAGCAGCAGCAGAAAGUGAAAUGUGAAAUGUUGCAACAAAAGGG ((((.............((((((....))))))......((((((((((((((..((-------------(((.((........)).)))))..).))))))))))))).......)))) ( -33.00) >DroYak_CAF1 14018 120 - 1 CCCUAAGAACGCACACUAUUUGCUUAUGCAGAUUUCGACCAACAUUUCGCAUUUGUUUUUUUUUUUUUUUUUUGGCAAACAGCGGCAGAAAGUGAAAUGUGAAAUGUUGCAACAAAAGGG ((((.............((((((....))))))......(((((((((((((((.............(((((((.(.......).)))))))..))))))))))))))).......)))) ( -28.96) >consensus CCCUAAGAACGCACACUAUUUGCUUAUGCAGAUUUCGACCAACAUUUCGCAUUUGUUUUC_________AAACGGCAAACAGCAGCAGAAAGUGAAAUGUGAAAUGUUGCAACAAAAGGG ((((.............((((((....))))))......(((((((((((((((....................((.....)).((.....)).))))))))))))))).......)))) (-26.60 = -26.80 + 0.20)


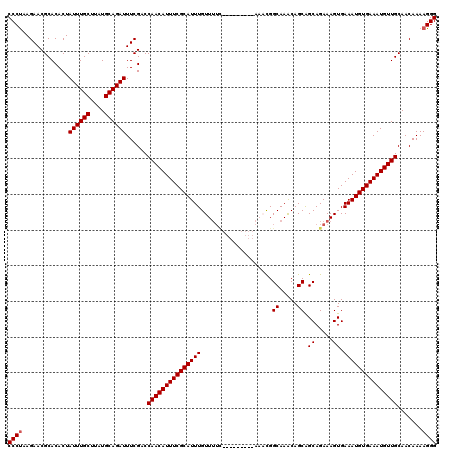
| Location | 19,883,869 – 19,883,984 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -22.36 |
| Consensus MFE | -18.24 |
| Energy contribution | -18.08 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19883869 115 + 22224390 GGUCGAAAUCUGCAUAAGCAAAUAGUGUGCGUUCUUAGGGGAUGGCACUUUCCCAUGCCCCAUGUCCCCAUGUCCCCAUGUCCCCAUUUCACCAUAUCACC---AUUUU--CCCAAUUCC (((.(((((..(((((.........))))).......((((((((((........)))).((((....)))).......))))))))))))))........---.....--......... ( -26.70) >DroSec_CAF1 37612 92 + 1 GGUCGAAAUCUGCAUAAGCAAAUAGUGUGCGUUCUUGGGGGAUGGCACUUUCCC-----------------------AUGUCCCCAUUUCACCAUAUCACC---AUUUC--CCCAUUUAC ((..(((((.(((....)))....(((((.((...((((((((((.......))-----------------------)).))))))....)))))))....---)))))--.))...... ( -21.90) >DroSim_CAF1 28357 92 + 1 GGUCGAAAUCUGCAUAAGCAAAUAGUGUGCGUUCUUAGGGGAUGGCACUUUCCC-----------------------AUGUCCCCAUUUCACCAUAUCACC---AUUUC--CCCAUUUAC (((.(((((..(((((.........))))).......((((((((.......))-----------------------)..)))))))))))))........---.....--......... ( -20.20) >DroEre_CAF1 17430 91 + 1 GGUCGAAAUCUGCAUAAGCAAAUAGUGUGCGUUCUUAGGGGAUAUCAC-UUCCC-----------------------AUGUCCCCAUUUCACCAUUUCCCC---AUUUC--CCCAUUUCC (((.(((((..(((((.........))))).......((((((((...-.....-----------------------))))))))))))))))........---.....--......... ( -21.80) >DroYak_CAF1 14098 97 + 1 GGUCGAAAUCUGCAUAAGCAAAUAGUGUGCGUUCUUAGGGGAUGGCACAUUCCC-----------------------AUGUCCCCAUUUCACCAUUUCCCCCAGAUUCCCACCCAUUUCC (((.(.((((((.....(((.......))).......((((((((.......))-----------------------)..)))))................)))))).).)))....... ( -21.20) >consensus GGUCGAAAUCUGCAUAAGCAAAUAGUGUGCGUUCUUAGGGGAUGGCACUUUCCC_______________________AUGUCCCCAUUUCACCAUAUCACC___AUUUC__CCCAUUUCC (((.(((((..(((((.........))))).......(((((((..................................)))))))))))))))........................... (-18.24 = -18.08 + -0.16)

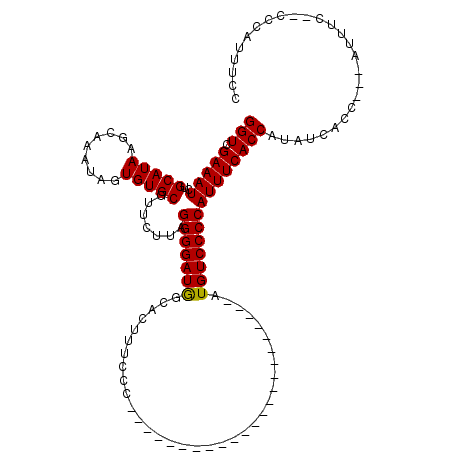
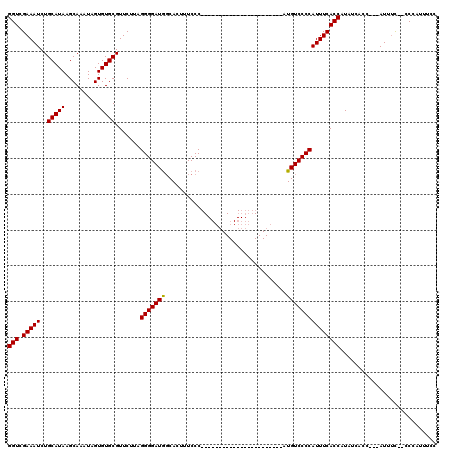
| Location | 19,883,869 – 19,883,984 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -29.78 |
| Consensus MFE | -17.74 |
| Energy contribution | -17.78 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19883869 115 - 22224390 GGAAUUGGG--AAAAU---GGUGAUAUGGUGAAAUGGGGACAUGGGGACAUGGGGACAUGGGGCAUGGGAAAGUGCCAUCCCCUAAGAACGCACACUAUUUGCUUAUGCAGAUUUCGACC ((((((...--.((((---((((.....(((..(((....)))(((((((((....)))).(((((......))))).)))))......))).))))))))((....)).)))))).... ( -40.90) >DroSec_CAF1 37612 92 - 1 GUAAAUGGG--GAAAU---GGUGAUAUGGUGAAAUGGGGACAU-----------------------GGGAAAGUGCCAUCCCCCAAGAACGCACACUAUUUGCUUAUGCAGAUUUCGACC ....(((((--.((((---((((.....(((...(((((..((-----------------------((.......)))).)))))....))).)))))))).)))))............. ( -25.10) >DroSim_CAF1 28357 92 - 1 GUAAAUGGG--GAAAU---GGUGAUAUGGUGAAAUGGGGACAU-----------------------GGGAAAGUGCCAUCCCCUAAGAACGCACACUAUUUGCUUAUGCAGAUUUCGACC ....(((((--.((((---((((...((((.....(((((..(-----------------------((.......)))))))).....)).)))))))))).)))))............. ( -24.60) >DroEre_CAF1 17430 91 - 1 GGAAAUGGG--GAAAU---GGGGAAAUGGUGAAAUGGGGACAU-----------------------GGGAA-GUGAUAUCCCCUAAGAACGCACACUAUUUGCUUAUGCAGAUUUCGACC .(((((..(--...((---(((.((((((((..(((....)))-----------------------((((.-......))))...........)))))))).))))).)..))))).... ( -25.00) >DroYak_CAF1 14098 97 - 1 GGAAAUGGGUGGGAAUCUGGGGGAAAUGGUGAAAUGGGGACAU-----------------------GGGAAUGUGCCAUCCCCUAAGAACGCACACUAUUUGCUUAUGCAGAUUUCGACC .......(((.(((((((((((.((((((((....(((((..(-----------------------((.......))))))))...(....).)))))))).)))...)))))))).))) ( -33.30) >consensus GGAAAUGGG__GAAAU___GGUGAUAUGGUGAAAUGGGGACAU_______________________GGGAAAGUGCCAUCCCCUAAGAACGCACACUAUUUGCUUAUGCAGAUUUCGACC ......((...((((...........(((((..(((....))).......................((((........))))...........)))))(((((....)))))))))..)) (-17.74 = -17.78 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:37 2006