
| Sequence ID | X_DroMel_CAF1 |
|---|---|
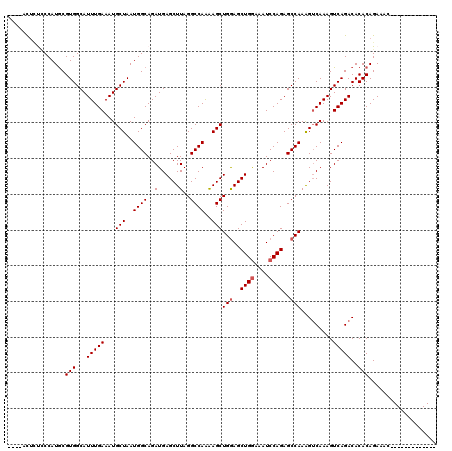
| Location | 19,883,318 – 19,883,573 |
| Length | 255 |
| Max. P | 0.961970 |

| Location | 19,883,318 – 19,883,429 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -28.75 |
| Consensus MFE | -21.20 |
| Energy contribution | -21.96 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19883318 111 + 22224390 ACUGACAAGAUAUAUGAUGGCAAACUUUAGCCAGCUUGCGUUGCUUUUUGUCGUUUGCCGCUCGCAGGCAAAUGGCAAAAGUUUGAUGU-----CCCCCCAA-CAACCAUAU---CCCCA ........(((((....((((........))))....(((((((((((.((((((((((.......))))))))))))))))..)))))-----........-.....))))---).... ( -30.00) >DroSec_CAF1 37072 102 + 1 ACUGACAAGAUAUAUGAUGGCAAACUUUAGCCAGCUU------------GUCGUUUGCCGCUCGCAGGCAAAUGGCAAAAGUUUGAUGCCACCUCCACCCAA-CGA-----CCACCCCCC ...((((((........((((........)))).)))------------)))(((((((.......)))))))((((.........))))............-...-----......... ( -25.40) >DroSim_CAF1 27798 120 + 1 ACUGACAAGAUAUAUGAUGGCAAACUUUAGCCAGCUUGCGUUGCUUUUUGUCGUUUGCCGCUCGCAGGCAAAUGGCAAAAGUUUGAUGCCACCUCCCCCCAAACAACCCCUCCUCCCCCC ...((..((........((((........))))....(((((((((((.((((((((((.......))))))))))))))))..)))))....................))..))..... ( -29.20) >DroEre_CAF1 16892 103 + 1 ACUGACAAGAUAUAUGAUGGCAAACUUUUGCCAGCUUGCGUUGCUUUUUGUCGUUGGCCGCUCGCAGGCAAAUGGUUAAAGUUUGAUAU-----CCCC-CAA-CACUCAC---------- ...............((((.((((((((.(((((((((((..(((..........)))....)))))))...)))).)))))))).)))-----)...-...-.......---------- ( -28.20) >DroYak_CAF1 13543 98 + 1 ACUGACAAGAUAUAUGAUGGCAAACUUUUGCCAGCUUGCGUUGCUUUUUGUCGUUUGCCGCUCGCAGGCAAAUGGUAAAAGUUUGAUAU-----CCCU-CCA-CU--------------- ...............((((.((((((((((((((((((((..((...............)).)))))))...))))))))))))).)))-----)...-...-..--------------- ( -30.96) >consensus ACUGACAAGAUAUAUGAUGGCAAACUUUAGCCAGCUUGCGUUGCUUUUUGUCGUUUGCCGCUCGCAGGCAAAUGGCAAAAGUUUGAUGU_____CCCCCCAA_CAA_C_______CCCC_ ....................((((((((.(((((((((((..((...............)).)))))))...)))).))))))))................................... (-21.20 = -21.96 + 0.76)

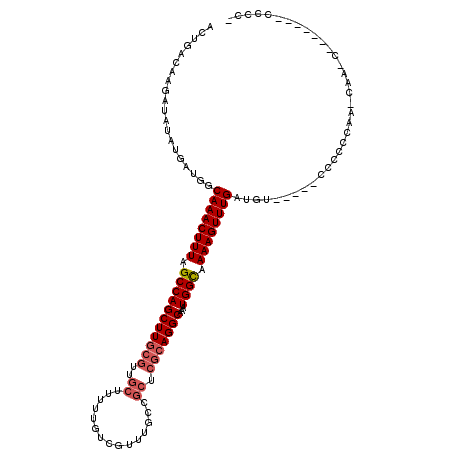
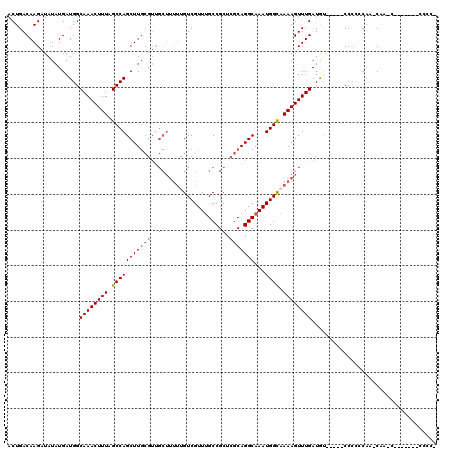
| Location | 19,883,358 – 19,883,469 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.39 |
| Mean single sequence MFE | -26.99 |
| Consensus MFE | -21.38 |
| Energy contribution | -22.54 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19883358 111 + 22224390 UUGCUUUUUGUCGUUUGCCGCUCGCAGGCAAAUGGCAAAAGUUUGAUGU-----CCCCCCAA-CAACCAUAU---CCCCACACUACUAUCCCAUGCGUGGCAUUUGAAAUGCUAAUGGCA .....((((((((((((((.......))))))))))))))(.(((.((.-----.....)).-))).)....---...............((((...((((((.....)))))))))).. ( -26.80) >DroSec_CAF1 37109 105 + 1 ---------GUCGUUUGCCGCUCGCAGGCAAAUGGCAAAAGUUUGAUGCCACCUCCACCCAA-CGA-----CCACCCCCCUCCCACUUUCCCAUACGUGGCAUUUGAAAUGCUAAUGGCA ---------((((((((((.......)))))))))).....((.((((((((..........-...-----.........................)))))))).))..(((.....))) ( -25.46) >DroSim_CAF1 27838 120 + 1 UUGCUUUUUGUCGUUUGCCGCUCGCAGGCAAAUGGCAAAAGUUUGAUGCCACCUCCCCCCAAACAACCCCUCCUCCCCCCUCGCACUUUCCCAUGCGUGGCAUUUGAAAUGCUAAUGGCA ....(((((((((((((((.......))))))))))))))).....(((((..............................((((........))))((((((.....)))))).))))) ( -33.70) >DroEre_CAF1 16932 99 + 1 UUGCUUUUUGUCGUUGGCCGCUCGCAGGCAAAUGGUUAAAGUUUGAUAU-----CCCC-CAA-CACUCAC--------------UUGCCACCAUGCGUGGCAUUUGAAAUGCUAAUGGCA ........((((((((((.(((....)))....................-----....-...-...(((.--------------.((((((.....))))))..)))...)))))))))) ( -26.20) >DroYak_CAF1 13583 94 + 1 UUGCUUUUUGUCGUUUGCCGCUCGCAGGCAAAUGGUAAAAGUUUGAUAU-----CCCU-CCA-CU-------------------UCUCCUCCAUGCGUGGCAUUUGAAAUGCUAAUGGCA ....(((((..((((((((.......))))))))..)))))........-----....-...-..-------------------.........(((.((((((.....))))))...))) ( -22.80) >consensus UUGCUUUUUGUCGUUUGCCGCUCGCAGGCAAAUGGCAAAAGUUUGAUGU_____CCCCCCAA_CAA_C_______CCCC_____ACUCUCCCAUGCGUGGCAUUUGAAAUGCUAAUGGCA ....(((((((((((((((.......))))))))))))))).................................................((((...((((((.....)))))))))).. (-21.38 = -22.54 + 1.16)


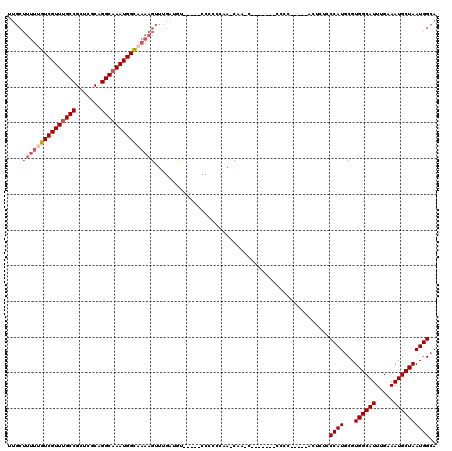
| Location | 19,883,429 – 19,883,536 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.31 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -23.82 |
| Energy contribution | -24.22 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19883429 107 + 22224390 CACUACUAUCCCAUGCGUGGCAUUUGAAAUGCUAAUGGCAGAUGAGCUUAGGCCAAAAGCUGAAGCUGGAAAUCCAGAGCCAAAGUCAAAGUCAGACACACAGAAAC------------- ................(((((((.....))))...((((.....(((((.(((.....))).)))))((....))...))))..(((.......))).)))......------------- ( -24.20) >DroSec_CAF1 37174 107 + 1 UCCCACUUUCCCAUACGUGGCAUUUGAAAUGCUAAUGGCAGAUGAGCUUAGGCCAAGAGCUGGAGCUGGAAAUCCAGAGCCAAAGUCAAAGUCAGACACACAGAAAC------------- .................((((.(((((...(((..((((..(......)..))))..)))(((..((((....))))..)))...))))))))).............------------- ( -27.70) >DroSim_CAF1 27918 107 + 1 UCGCACUUUCCCAUGCGUGGCAUUUGAAAUGCUAAUGGCAGAUGAGCUUAGGCCAAGAGCUGGAGCUGGAAAUCCAGAGCCAAGGACAAAGUCAGACACACAGAAAC------------- .((((........))))((((.((((....(((..((((..(......)..))))..)))(((..((((....))))..)))....)))))))).............------------- ( -29.50) >DroEre_CAF1 16995 103 + 1 ----UUGCCACCAUGCGUGGCAUUUGAAAUGCUAAUGGCAGAUGAGCUUAGGCCAAAAGCUGGAGCUGGAAAUCCAGAGCCAAAGUCAAAGUCAGACACACAGAAAC------------- ----.((((((.....))))))((((....(((..((((..(......)..))))..)))(((..((((....))))..)))..(((.......)))...))))...------------- ( -31.50) >DroYak_CAF1 13641 116 + 1 ----UCUCCUCCAUGCGUGGCAUUUGAAAUGCUAAUGGCAGAUGAGCUUAGGCCAGAAGCUGGAGCUGAAAAUCCAGAGCCAAAGUCAAAGUCAGACACACAGAAAGUCGAAGGCUCGAA ----((..(((((.((.((((((.....)))))).((((..(......)..))))...)))))))..)).......(((((...(((.......))).....(.....)...)))))... ( -32.20) >consensus ____ACUCUCCCAUGCGUGGCAUUUGAAAUGCUAAUGGCAGAUGAGCUUAGGCCAAAAGCUGGAGCUGGAAAUCCAGAGCCAAAGUCAAAGUCAGACACACAGAAAC_____________ ................(((...(((((...(((..((((..(......)..))))..)))(((..((((....))))..))).........)))))..)))................... (-23.82 = -24.22 + 0.40)

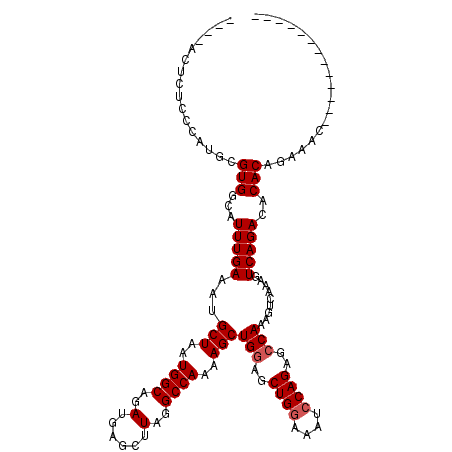
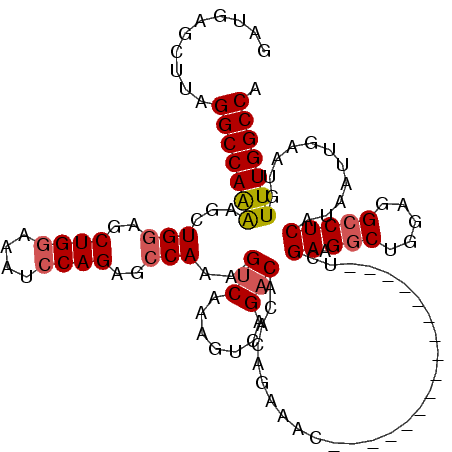
| Location | 19,883,469 – 19,883,573 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.76 |
| Mean single sequence MFE | -31.16 |
| Consensus MFE | -24.56 |
| Energy contribution | -24.96 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19883469 104 + 22224390 GAUGAGCUUAGGCCAAAAGCUGAAGCUGGAAAUCCAGAGCCAAAGUCAAAGUCAGACACACAGAAAC----------------UUGAAGGCUGGAGGCCUCAUAAUUGAAUGUUUGGCCA ..(((.(((.((((...(((....)))((....)).).))).))))))..((((((((..(((....----------------.(((.(((.....))))))...)))..)))))))).. ( -28.30) >DroSec_CAF1 37214 104 + 1 GAUGAGCUUAGGCCAAGAGCUGGAGCUGGAAAUCCAGAGCCAAAGUCAAAGUCAGACACACAGAAAC----------------UCGAAGGCUGGAGGCCUCAUAAUUGAAUGUUUGGCCA ..........(((((((((.(((..((((....))))..)))..(((.......))).........)----------------))((.(((.....)))))............)))))). ( -32.30) >DroSim_CAF1 27958 104 + 1 GAUGAGCUUAGGCCAAGAGCUGGAGCUGGAAAUCCAGAGCCAAGGACAAAGUCAGACACACAGAAAC----------------UCGAAGGCUGGAGGCCUCAUAAUUGAAUGUUUGGCCA ..........(((((((((.(((..((((....))))..)))..(((...))).............)----------------))((.(((.....)))))............)))))). ( -30.80) >DroEre_CAF1 17031 104 + 1 GAUGAGCUUAGGCCAAAAGCUGGAGCUGGAAAUCCAGAGCCAAAGUCAAAGUCAGACACACAGAAAC----------------UCGAAGGCUGGAGGCCUCAUAAUUGAAUGUUUGGCCA ..........(((((((...(((..((((....))))..)))..(((.......)))..........----------------..((.(((.....)))))...........))))))). ( -30.10) >DroYak_CAF1 13677 120 + 1 GAUGAGCUUAGGCCAGAAGCUGGAGCUGAAAAUCCAGAGCCAAAGUCAAAGUCAGACACACAGAAAGUCGAAGGCUCGAAGACUCGAAGACUGGAGGCCUCAUAAUUGAAUGUUUGGCCA ..........(((((((..(((((........))))).(((..((((...(((.(((.........)))...)))((((....)))).))))...)))..............))))))). ( -34.30) >consensus GAUGAGCUUAGGCCAAAAGCUGGAGCUGGAAAUCCAGAGCCAAAGUCAAAGUCAGACACACAGAAAC________________UCGAAGGCUGGAGGCCUCAUAAUUGAAUGUUUGGCCA ..........(((((((...(((..((((....))))..)))..(((.......)))............................((.(((.....)))))...........))))))). (-24.56 = -24.96 + 0.40)


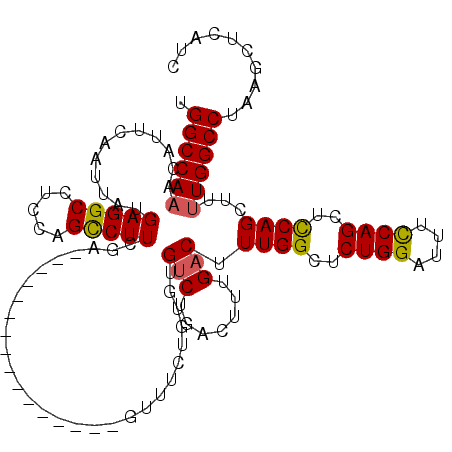
| Location | 19,883,469 – 19,883,573 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.76 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -22.92 |
| Energy contribution | -22.84 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.783846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19883469 104 - 22224390 UGGCCAAACAUUCAAUUAUGAGGCCUCCAGCCUUCAA----------------GUUUCUGUGUGUCUGACUUUGACUUUGGCUCUGGAUUUCCAGCUUCAGCUUUUGGCCUAAGCUCAUC .((((((((((..((((..(((((.....)))))..)----------------)))...))))(((.......)))...((((((((....))))....)))).)))))).......... ( -27.40) >DroSec_CAF1 37214 104 - 1 UGGCCAAACAUUCAAUUAUGAGGCCUCCAGCCUUCGA----------------GUUUCUGUGUGUCUGACUUUGACUUUGGCUCUGGAUUUCCAGCUCCAGCUCUUGGCCUAAGCUCAUC .((((((............(((((.....))))).((----------------((........(((.......)))..(((..((((....))))..))))))))))))).......... ( -29.80) >DroSim_CAF1 27958 104 - 1 UGGCCAAACAUUCAAUUAUGAGGCCUCCAGCCUUCGA----------------GUUUCUGUGUGUCUGACUUUGUCCUUGGCUCUGGAUUUCCAGCUCCAGCUCUUGGCCUAAGCUCAUC .((((((((((..((((..(((((.....)))))..)----------------)))...))))..............((((..((((....))))..))))...)))))).......... ( -28.20) >DroEre_CAF1 17031 104 - 1 UGGCCAAACAUUCAAUUAUGAGGCCUCCAGCCUUCGA----------------GUUUCUGUGUGUCUGACUUUGACUUUGGCUCUGGAUUUCCAGCUCCAGCUUUUGGCCUAAGCUCAUC .((((((((((..((((..(((((.....)))))..)----------------)))...))))(((.......))).((((..((((....))))..))))...)))))).......... ( -29.90) >DroYak_CAF1 13677 120 - 1 UGGCCAAACAUUCAAUUAUGAGGCCUCCAGUCUUCGAGUCUUCGAGCCUUCGACUUUCUGUGUGUCUGACUUUGACUUUGGCUCUGGAUUUUCAGCUCCAGCUUCUGGCCUAAGCUCAUC .(((((...............((((...((((...(((((...(......)(((.........))).))))).))))..))))(((((........)))))....))))).......... ( -29.60) >consensus UGGCCAAACAUUCAAUUAUGAGGCCUCCAGCCUUCGA________________GUUUCUGUGUGUCUGACUUUGACUUUGGCUCUGGAUUUCCAGCUCCAGCUUUUGGCCUAAGCUCAUC .((((((............(((((.....))))).............................(((.......))).((((..((((....))))..))))...)))))).......... (-22.92 = -22.84 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:32 2006