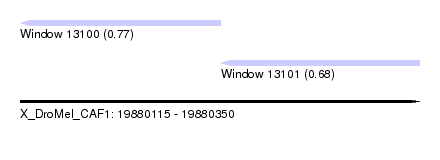
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,880,115 – 19,880,350 |
| Length | 235 |
| Max. P | 0.767634 |

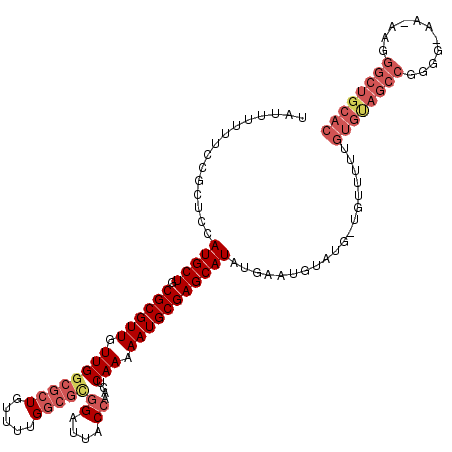
| Location | 19,880,115 – 19,880,233 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.19 |
| Mean single sequence MFE | -37.98 |
| Consensus MFE | -22.76 |
| Energy contribution | -25.83 |
| Covariance contribution | 3.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19880115 118 - 22224390 UAUUUUUUCCGCUCCAUGCUGCGCGUUGUUGGCGCUGUUUUGGCGUGGAUUACCAACUCAAAAAUGCGAGCAUAUGAAUGUUUG-UGUUUUUGUGUAGCCGUGGGAAAAAGGGCUGCAC .......(((((.(((.((.((((.......)))).))..))).))))).........(((((((((((((((....)))))))-))))))))(((((((.(.......).))))))). ( -44.60) >DroSec_CAF1 33958 118 - 1 UAUUUUUUCCGCUCCAUGCUGCGCGUUGUUGGCGCUGUUUUGGCGCGGAUUACGAACUCAAAAAUGCGAGCAUAUGAAUGCAUGUUGUUUUUGUGUAGCCGAGGGAA-AAGGGCUGCAC ..((((((((.(((...(((((.(((.(((.(((((.....))))).))).)))....((((((((((.((((....)))).)).)))))))).))))).)))))))-))))....... ( -39.80) >DroEre_CAF1 11276 104 - 1 UAUUU-UUCCGCUCCAUGCUGCGCGUUGUUG-----------GCGCGGAUUACCAACUCAAAAAUGCGAGCAUAUGUAUGUAUG-UGUUUUUGUGCAGCCGGGG-GA-AAGGGCUGCAC .....-.(((((.(((.((........))))-----------).))))).........((((((((((.((((....)))).))-))))))))(((((((....-..-...))))))). ( -35.00) >DroYak_CAF1 9687 97 - 1 UAUUUUUUCCGCUCCAUGCUGCGCGUUGUUGGCGCUGUUUUGGCGCGGAUUACCAACUCAAAAAUGCGAGCAUAUGUACG--------------------AGGG-AA-AAGGGCUGCAC ..((((((((.(((.(((((.((((((.((((((((.....)))))((....))....))).)))))))))))......)--------------------))))-))-))))....... ( -32.50) >consensus UAUUUUUUCCGCUCCAUGCUGCGCGUUGUUGGCGCUGUUUUGGCGCGGAUUACCAACUCAAAAAUGCGAGCAUAUGAAUGUAUG_UGUUUUUGUGUAGCCGGGG_AA_AAGGGCUGCAC ...............(((((.((((((.((((((((.....)))))((....))....))).)))))))))))...................((((((((...........)))))))) (-22.76 = -25.83 + 3.06)

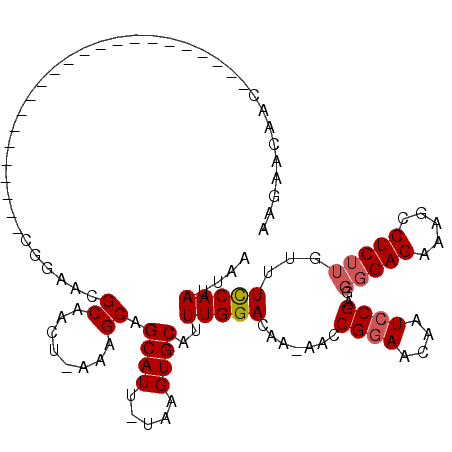
| Location | 19,880,233 – 19,880,350 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.39 |
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -13.76 |
| Energy contribution | -14.33 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19880233 117 - 22224390 AAGAACAACAAUAACAUAAAUGUACGCCCUUCCGGAACGCAACUUAAAGCAGCAUU-UAAGUGCAUUUGGACAA-AACCGGAACAAUCCGA-GGGGAGAAAGCCUCUUGUUUCCAAUUAA ..((((((.....(((....))).(.((((((.(((..(((.((((((......))-))))))).(((((....-..)))))....)))))-)))).)........))))))........ ( -26.50) >DroSec_CAF1 34076 113 - 1 AAGAACAACA---ACAGAAAUGUACCCCCUUCCGGAACGCAACU-AAAGCAGCAUU-UAAGUGCAUUUGGACAA-AACCGGAACGAUCCGA-GGGGAGAAAGCCUCUUGUUUCCAAUUAA ..((((((..---(((....)))..(((((((((((..((....-...)).((((.-...)))).))))))...-...((((....)))))-))))..........))))))........ ( -28.00) >DroEre_CAF1 11380 91 - 1 AACAACGGG-----------------------GGGAACGCAACU-AAAGCAGCAUUUUAAGUGCAUUUGGACAA-AACCGAAACAAUCCGA----GAGAAAGCCUCUUUUUUCCAAUUAA ......(((-----------------------((((..((....-...)).((((.....)))).(((((....-..)))))....)))((----(((.....)))))..))))...... ( -17.90) >DroYak_CAF1 9784 96 - 1 AAGAGCAAC-----------------------UGAAAUGCAACU-AAGGCAGCAUUUUAAGUGCAUUUGGACAAAAACCGGAACAAUCCGAAGGGGAGAAAGCCUCUUGUUUUCAAUUAA ....(((..-----------------------((((((((..(.-...)..))))))))..)))..(((((.......((((....))))((((((......))))))...))))).... ( -24.10) >consensus AAGAACAAC_______________________CGGAACGCAACU_AAAGCAGCAUU_UAAGUGCAUUUGGACAA_AACCGGAACAAUCCGA_GGGGAGAAAGCCUCUUGUUUCCAAUUAA ......................................((........)).((((.....))))..(((((.......((((....))))...(((((.....)))))...))))).... (-13.76 = -14.33 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:25 2006