
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,879,106 – 19,879,284 |
| Length | 178 |
| Max. P | 0.554937 |

| Location | 19,879,106 – 19,879,208 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 58.53 |
| Mean single sequence MFE | -23.27 |
| Consensus MFE | -13.00 |
| Energy contribution | -13.67 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
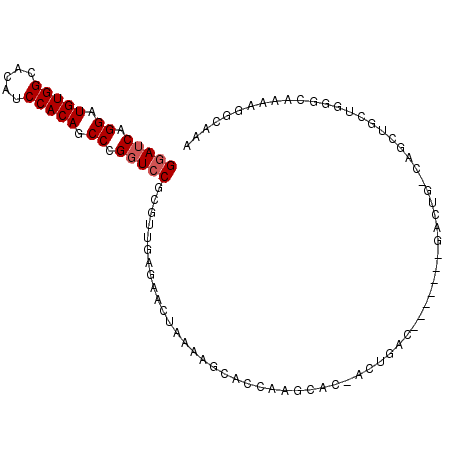
>X_DroMel_CAF1 19879106 102 + 22224390 GGAUCAGGAUGUGGCACAUCCACAACCCGUUCCGCUUUGAGAAGCACUAGCACCAAGCACUACUGACGACUGACGACUGACAGCUGCUGGGCAAAAGGCAAA (((.(.((.(((((.....))))).)).).)))(((((.....((.((((((.((........))..(.(((........))))))))))))..)))))... ( -26.80) >DroSec_CAF1 33030 95 + 1 GGAUCAGGAUGUGGCACAUCCACAGCCCGGUACUCGUUGAGAACUAAAAGCACCAAGCACCACUGAC-------GACUGGCAGCUGCUGGGCAAAAGGCAAA ......(((((.....)))))...((((((((...((((..........((.....)).(((.....-------...))))))))))))))).......... ( -26.30) >DroSim_CAF1 25380 95 + 1 GGAUCAGGAUGUGGCACAUCCACAGCCCGGUCCGCGUUGAGAACUAAAACCNNNNNNNNNNNNNNNN-------NNNNNNNNNNNNNNNNNNNNNNNNNNNN (((((.((.(((((.....))))).)).)))))..................................-------............................ ( -16.70) >consensus GGAUCAGGAUGUGGCACAUCCACAGCCCGGUCCGCGUUGAGAACUAAAAGCACCAAGCAC_ACUGAC_______GACUG_CAGCUGCUGGGCAAAAGGCAAA (((((.((.(((((.....))))).)).)))))..................................................................... (-13.00 = -13.67 + 0.67)


| Location | 19,879,185 – 19,879,284 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.67 |
| Mean single sequence MFE | -35.86 |
| Consensus MFE | -25.90 |
| Energy contribution | -26.03 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
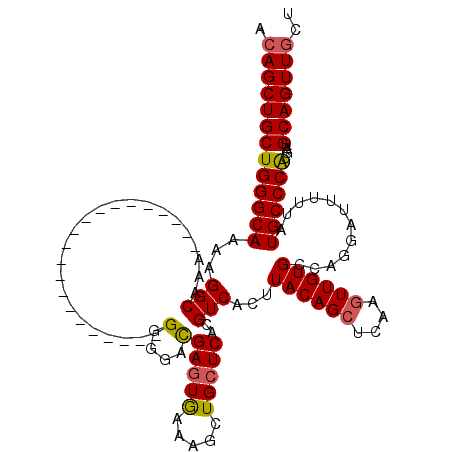
>X_DroMel_CAF1 19879185 99 + 22224390 ACAGCUGCUGGGCAAAAGGCAAA---------------------GGGGUCGAGUGAAAGCUGGUCACGUCACUUACAGCUCAAGUUGUGCCAGGAUUUUUAUGCCCGCUAUGCAGUUGCU .(((((((..(((....((((.(---------------------((((((.(((....)))((.((((..((((.......))))))))))..))))))).)))).)))..))))))).. ( -32.60) >DroSec_CAF1 33102 99 + 1 GCAGCUGCUGGGCAAAAGGCAAA---------------------GGGGUCGAGUGAAAGCUGCUCACGUCACUUACAGCUCAAGUUGUGCCAGGAUUUUUAUGCCCGCGAUGCAGUUGCU ((((((((((((((...(((...---------------------.((((..(((....)))))))..)))...((((((....))))))............))))))....)))))))). ( -35.60) >DroEre_CAF1 10417 120 + 1 ACAGCUGCUGGGCAAACGGCAAAGGGCAAACGGCAAACGGCAAAGGGGAUGAGUAAAAGCUGCUCACGUCACUUACAGCUCAACUUGUGCCAGGAUUUUUAUGCCCACGAUGCAGUUGCU .(((((((((((((....((.....((.....)).....))....((.(..(((...(((((.............)))))..)))..).))..........))))))....))))))).. ( -36.92) >DroYak_CAF1 8718 111 + 1 ACAGCUGCUGGGCAAAUGGCAAA-----AAGGGCAAA----AUGGGGGACGAGUGCAAGCUGCUCACGUCACUUACAGCUCAAGUUGUGCCAGGAUUUGUAUGCCCACGAUGCAGUUCCU ..((((((((((((....(((((-----...((((..----.((((.((((.(.((.....)).).)))).))))((((....))))))))....))))).))))))....))))))... ( -38.30) >consensus ACAGCUGCUGGGCAAAAGGCAAA_____________________GGGGACGAGUGAAAGCUGCUCACGUCACUUACAGCUCAAGUUGUGCCAGGAUUUUUAUGCCCACGAUGCAGUUGCU .(((((((((((((...(((........................(....)(((((.....)))))..)))...((((((....))))))............))))))....))))))).. (-25.90 = -26.03 + 0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:21 2006