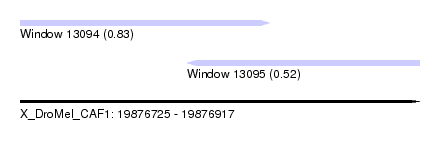
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,876,725 – 19,876,917 |
| Length | 192 |
| Max. P | 0.825196 |

| Location | 19,876,725 – 19,876,845 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.11 |
| Mean single sequence MFE | -16.23 |
| Consensus MFE | -14.86 |
| Energy contribution | -14.86 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.825196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
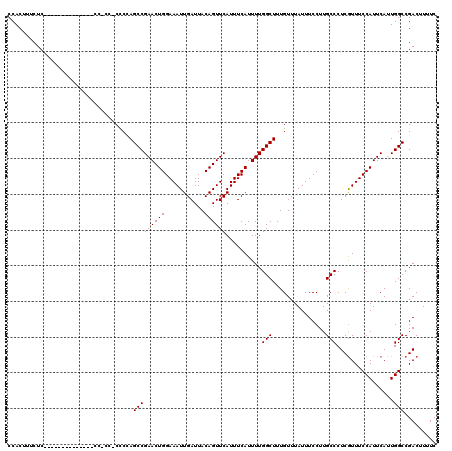
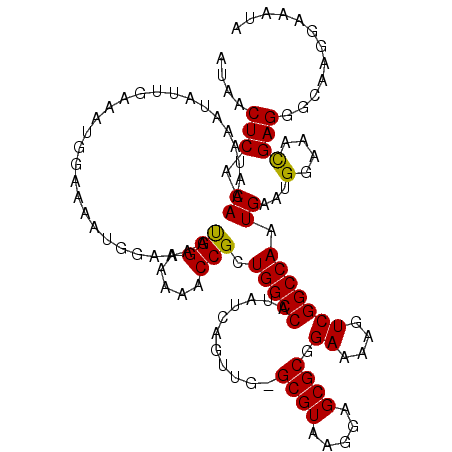
>X_DroMel_CAF1 19876725 120 + 22224390 CCACUUUCUCGCAUCCCCCUCCCCCCGCCCCUCUAGCCGAACUGGAAAUUGAUUACAGUUCAUUUCAUUUUGGCUUUGUUUAUUUCCUUGCCCUCGUUUCCAUUCAUUGGCCGACUUUUC ..................................(((((((.(((((..(((.......)))))))).)))))))..................(((...(((.....))).)))...... ( -15.30) >DroSec_CAF1 30778 100 + 1 CCACUUUCUC--------------------CCCCAGCCGAACUGGAAAUUGAUUACAGUUCAUUUCAUUUUGGCUUUGUUUAUUUCCUUGCCCUCGUUUCCAUUCAUUGGCCGACUUUUC ..........--------------------....(((((((.(((((..(((.......)))))))).)))))))..................(((...(((.....))).)))...... ( -15.40) >DroSim_CAF1 23078 100 + 1 CCACUUUCUC--------------------CCCCAGCCGAACUGGAAAUUGAUUACAGUUCAUUUCAUUUUGGCUUUGUUUAUUUCCUUGCCCUCGUUUCCAUUCAUUGGCCGACUUUUC ..........--------------------....(((((((.(((((..(((.......)))))))).)))))))..................(((...(((.....))).)))...... ( -15.40) >DroEre_CAF1 8162 105 + 1 CCACUUCCUC--------------CCACCGCCC-AGCCGAACUGGAAAUUGAUUACAGUUCAUUUCAUUUUGGCUUUGUUUAUUUCCUUGCCCUCAUUUCCAUUCACUGGCCGACUUUUC ..........--------------....((.((-((..(((.((((((((((.......))).........(((...............)))...)))))))))).)))).))....... ( -18.36) >DroYak_CAF1 6253 106 + 1 CCACUGCCUC--------------CCACCCCGCAAGCCGAACUGGAAAUUGAUUACAGUUCAUUUCAUUUUGGCUUUGUUUAUUUCCUUGCCCUCGUUUCCAUUCACUGGCCGACUUUUC ...(.(((..--------------.......((((((((((.(((((..(((.......)))))))).)))))).))))................((........)).))).)....... ( -16.70) >consensus CCACUUUCUC______________CC_CC_CCCCAGCCGAACUGGAAAUUGAUUACAGUUCAUUUCAUUUUGGCUUUGUUUAUUUCCUUGCCCUCGUUUCCAUUCAUUGGCCGACUUUUC ...................................(((((((((...........))))))..........(((...............)))................)))......... (-14.86 = -14.86 + 0.00)


| Location | 19,876,805 – 19,876,917 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.27 |
| Mean single sequence MFE | -23.28 |
| Consensus MFE | -18.50 |
| Energy contribution | -18.18 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19876805 112 - 22224390 AUAACUCACAAAUAAAUAUUGAAAUG-------GAAAAUGGAAAAACCGCUGGCCAUAUCAGUUG-GCGUAAGGAGCGCGGAAAAGUCGGCCAAUGAAUGGAAACGAGGGCAAGGAAAUA ....(((............(((.(((-------(..(.(((.....))).)..)))).)))((((-((((.....)).(((.....)))))))))....(....))))............ ( -21.60) >DroSec_CAF1 30838 112 - 1 AUAACUCACAAAUAAAUAUUAAAAUG-------GAAAAUGGAAAAACCGCUGGCCAUAUCAGUUG-GCGUAAGGAGCGCGGAAAAGUCGGCCAAUGAAUGGAAACGAGGGCAAGGAAAUA ....(((.................((-------(....(((.....((((..((((.......))-))((.....)))))).....))).)))......(....))))............ ( -21.30) >DroSim_CAF1 23138 119 - 1 AUAACUCACAAAUAAAUAUUGAAAUGGAAAAUGGAAAAUGGAAAAACCGCUGGCCAUAUCAGUUG-GCGUAAGGAGCGCGGAAAAGUCGGCCAAUGAAUGGAAACGAGGGCAAGGAAAUA ....(((........................(((....(((.....((((..((((.......))-))((.....)))))).....))).)))......(....))))............ ( -21.30) >DroEre_CAF1 8227 119 - 1 AUAACUCACAAAUAAAUAUUGAAAUGGAAAAUGGAAAAUGGAAAACCCGCUGGCCAUAUCAGUUG-GCGUAAGGAGCGCGGAAAAGUCGGCCAGUGAAUGGAAAUGAGGGCAAGGAAAUA ....((((...............................((....))((((((((.......((.-((((.....)))).))......))))))))........))))............ ( -25.82) >DroYak_CAF1 6319 120 - 1 AUAACUCACAAAUAAAUAUUGAAAUGGAAAAUGGAAAACGGAAAAACCGCUGGCCAUAUCAGUUGAGCGUAAGGAGCGCGGAAAAGUCGGCCAGUGAAUGGAAACGAGGGCAAGGAAAUA ....(((........................................((((((((...((....))((((.....)))).........))))))))...(....))))............ ( -26.40) >consensus AUAACUCACAAAUAAAUAUUGAAAUGGAAAAUGGAAAAUGGAAAAACCGCUGGCCAUAUCAGUUG_GCGUAAGGAGCGCGGAAAAGUCGGCCAAUGAAUGGAAACGAGGGCAAGGAAAUA ....(((.((............................(((.....))).(((((...........((((.....)))).((....))))))).))...(....))))............ (-18.50 = -18.18 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:20 2006