
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,873,412 – 19,873,572 |
| Length | 160 |
| Max. P | 0.861707 |

| Location | 19,873,412 – 19,873,532 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -41.61 |
| Consensus MFE | -41.12 |
| Energy contribution | -41.23 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
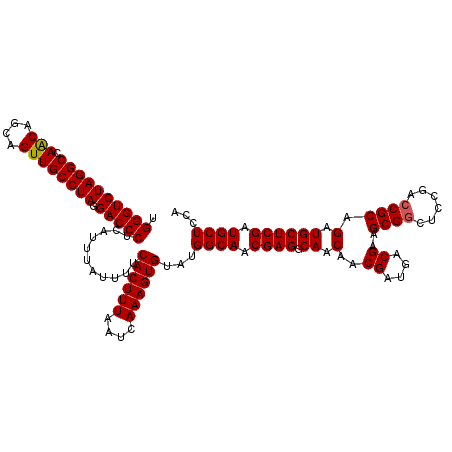
>X_DroMel_CAF1 19873412 120 + 22224390 UGGAGCAUCGAGCAUCUGCGGUCGGAGCCGCUUCGUCAUCGUUGUUGCCUCGUUGCCAUACACUUUGAUUAAAGUGAAAAUAAAUGAGGGUCCUAGGCAACUGCUGUUGGCCUAGACCCA .((((((.(((......(((((....))))).......))).)))).))(((((......((((((....))))))......)))))(((((.((((((((....))).)))))))))). ( -42.42) >DroSec_CAF1 27603 120 + 1 UGGAGCAUCGAGCAUCUGCGGUCGGAGCCGCUUCGUCAUCGUUGUUGCCUCGUUGCCAUACACUUUGAUUAAAGUGAAAAUAAAUGAGGGUCCUAGGCAACUGCUGUUGGCCUAGACCCA .((((((.(((......(((((....))))).......))).)))).))(((((......((((((....))))))......)))))(((((.((((((((....))).)))))))))). ( -42.42) >DroEre_CAF1 4993 120 + 1 UGGAGCAUCGAGCAUCUGCGGUCGAAGUCGCUUCGUCAUCGUUGUUGCCUCGUUGCCAUACACUUUGAUUAAAGUGAAAAUAAAUGAGGGUCCUAGGCAACUGCUGCUGGCCUAGACCCA ..(.(((.((((((...((((((((((...)))))..)))))...)).)))).))))...((((((....))))))...........(((((.(((((..(....)...)))))))))). ( -40.00) >consensus UGGAGCAUCGAGCAUCUGCGGUCGGAGCCGCUUCGUCAUCGUUGUUGCCUCGUUGCCAUACACUUUGAUUAAAGUGAAAAUAAAUGAGGGUCCUAGGCAACUGCUGUUGGCCUAGACCCA ..(.(((.((((((...((((((((((...)))))..)))))...)).)))).))))...((((((....))))))...........(((((.((((((((....))).)))))))))). (-41.12 = -41.23 + 0.11)


| Location | 19,873,412 – 19,873,532 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -37.97 |
| Consensus MFE | -37.42 |
| Energy contribution | -37.53 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19873412 120 - 22224390 UGGGUCUAGGCCAACAGCAGUUGCCUAGGACCCUCAUUUAUUUUCACUUUAAUCAAAGUGUAUGGCAACGAGGCAACAACGAUGACGAAGCGGCUCCGACCGCAGAUGCUCGAUGCUCCA .((((((((((.(((....)))))))).)))))...........((((((....))))))...((((.((((.((.(..((....))..((((......)))).).)))))).))))... ( -39.00) >DroSec_CAF1 27603 120 - 1 UGGGUCUAGGCCAACAGCAGUUGCCUAGGACCCUCAUUUAUUUUCACUUUAAUCAAAGUGUAUGGCAACGAGGCAACAACGAUGACGAAGCGGCUCCGACCGCAGAUGCUCGAUGCUCCA .((((((((((.(((....)))))))).)))))...........((((((....))))))...((((.((((.((.(..((....))..((((......)))).).)))))).))))... ( -39.00) >DroEre_CAF1 4993 120 - 1 UGGGUCUAGGCCAGCAGCAGUUGCCUAGGACCCUCAUUUAUUUUCACUUUAAUCAAAGUGUAUGGCAACGAGGCAACAACGAUGACGAAGCGACUUCGACCGCAGAUGCUCGAUGCUCCA .((((((((((.(((....)))))))).)))))...........((((((....))))))...((((.((((.((.(..((....(((((...)))))..))..).)))))).))))... ( -35.90) >consensus UGGGUCUAGGCCAACAGCAGUUGCCUAGGACCCUCAUUUAUUUUCACUUUAAUCAAAGUGUAUGGCAACGAGGCAACAACGAUGACGAAGCGGCUCCGACCGCAGAUGCUCGAUGCUCCA .((((((((((.(((....)))))))).)))))...........((((((....))))))...((((.((((.((.(..((....))..((((......)))).).)))))).))))... (-37.42 = -37.53 + 0.11)


| Location | 19,873,452 – 19,873,572 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.23 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -25.33 |
| Energy contribution | -25.67 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
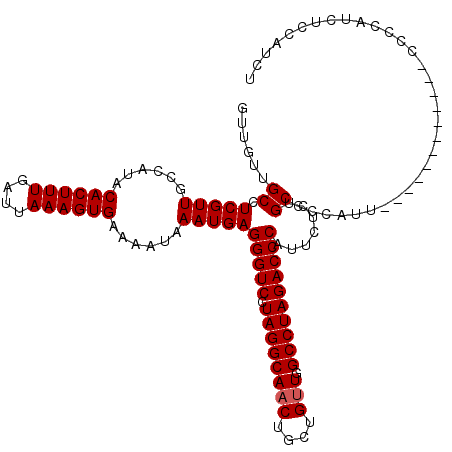
>X_DroMel_CAF1 19873452 120 + 22224390 GUUGUUGCCUCGUUGCCAUACACUUUGAUUAAAGUGAAAAUAAAUGAGGGUCCUAGGCAACUGCUGUUGGCCUAGACCCAUUCUCUGCCCCAUUCCCAAUCUCCAUCUCCAUCUCCACCU (..((((..(((((......((((((....))))))......)))))(((((.((((((((....))).)))))))))).................))))..)................. ( -26.80) >DroSec_CAF1 27643 108 + 1 GUUGUUGCCUCGUUGCCAUACACUUUGAUUAAAGUGAAAAUAAAUGAGGGUCCUAGGCAACUGCUGUUGGCCUAGACCCAUUCUCUGCCCCAUU------------CCCCAUCCCCAUCU ......((.(((((......((((((....))))))......)))))(((((.((((((((....))).)))))))))).......))......------------.............. ( -26.70) >DroEre_CAF1 5033 108 + 1 GUUGUUGCCUCGUUGCCAUACACUUUGAUUAAAGUGAAAAUAAAUGAGGGUCCUAGGCAACUGCUGCUGGCCUAGACCCAUUCUAUGCCCCAUU------------CCCCAUCUCCAUCU .......(((((((......((((((....))))))......)))))))((..(((....)))..)).(((.((((.....)))).))).....------------.............. ( -24.20) >consensus GUUGUUGCCUCGUUGCCAUACACUUUGAUUAAAGUGAAAAUAAAUGAGGGUCCUAGGCAACUGCUGUUGGCCUAGACCCAUUCUCUGCCCCAUU____________CCCCAUCUCCAUCU ......((.(((((......((((((....))))))......)))))(((((.((((((((....))).)))))))))).......))................................ (-25.33 = -25.67 + 0.33)


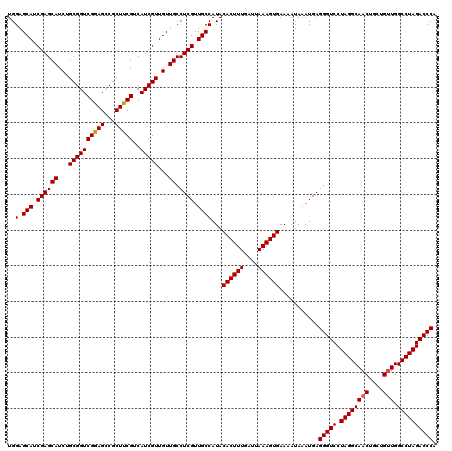
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:13 2006