
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,871,953 – 19,872,095 |
| Length | 142 |
| Max. P | 0.975274 |

| Location | 19,871,953 – 19,872,066 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.77 |
| Mean single sequence MFE | -35.34 |
| Consensus MFE | -31.04 |
| Energy contribution | -31.24 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19871953 113 + 22224390 UUCAAGCAAAACAAUUUUGCAUUACAAAAGGGCAUAGAGCUCACGGGCCGCUGACCACCGACCAGAAAGCGCAACAAAGUGCUGGACACUGGUCGCCCCACGGGAUUUUGAGG------- ((((((((((.....))))).........((((.(((.(((....)))..)))......((((((..(((((......))))).....)))))))))).........))))).------- ( -34.50) >DroSec_CAF1 26174 113 + 1 UUCAAGCAAAACAAUUUUGCAUUACAAAAGGGCAUAGAGCUCACGGGCCGCUGACCACCGACCAGAAAGCGCAACAAAGUGCUGGACACUGGUCGCCCCACGGGACUUUGCCG------- .....(((((.....)))))..........((((..((((((..((((....(.....)((((((..(((((......))))).....))))))))))...))).))))))).------- ( -35.30) >DroSim_CAF1 17314 119 + 1 UUCAAGCAAAACAAUUUUGCAUUACAAAAGGGCAUAGAGCUCACGGGCCGCUGAUCA-UGACCAGAAAGCGCAACAAAGUGCUGGACACUGGUCGCCCCACGGGACUUUGCCGGGAAAUG (((..(((((.....))))).........((((.(((.(((....)))..)))....-.((((((..(((((......))))).....))))))))))..(((.......))).)))... ( -35.30) >DroEre_CAF1 3722 112 + 1 UUCAAGCAAAACAAUUUUGCAUUACAAAAGGGCAUAGAGCUCACGGGCCGCUGACCACCGACCAGAAAGCGCAACAAAGUGCUGGACACUGGUCGCCCCGCGGGAUUUUCCC-------- .....(((((.....))))).........((((.(((.(((....)))..)))......((((((..(((((......))))).....))))))))))...(((.....)))-------- ( -35.70) >DroYak_CAF1 1254 115 + 1 UUCAAGCAAAACAAUUUUGCAUUACAAAAGGGCAUAGAGCUCACGGGCCGCUGACCAGCGACCAGAAAGCGCAACAAAGUGCUGGACACUGGUCACCCACAGGGAUUUUUCC-----ACG .....(((((.....))))).........((((.....))))..(((..(((....)))((((((..(((((......))))).....)))))).)))....(((....)))-----... ( -35.90) >consensus UUCAAGCAAAACAAUUUUGCAUUACAAAAGGGCAUAGAGCUCACGGGCCGCUGACCACCGACCAGAAAGCGCAACAAAGUGCUGGACACUGGUCGCCCCACGGGAUUUUGCCG_______ .....(((((.....))))).........((((.(((.(((....)))..)))......((((((..(((((......))))).....))))))))))...................... (-31.04 = -31.24 + 0.20)



| Location | 19,871,993 – 19,872,095 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -37.58 |
| Consensus MFE | -28.61 |
| Energy contribution | -28.61 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19871993 102 - 22224390 UUUU----------CCUGUGCUUUUCCCCGCAUUUCCCC--------CCUCAAAAUCCCGUGGGGCGACCAGUGUCCAGCACUUUGUUGCGCUUUCUGGUCGGUGGUCAGCGGCCCGUGA ...(----------(.((.(((.((..((((......((--------((.(........).))))(((((((.((.((((.....)))).))...)))))))))))..)).))).)).)) ( -34.00) >DroSec_CAF1 26214 111 - 1 UUUUCCCCUCAUUUCCCGCGUUUUUCCCCGCAUUUCCC---------CGGCAAAGUCCCGUGGGGCGACCAGUGUCCAGCACUUUGUUGCGCUUUCUGGUCGGUGGUCAGCGGCCCGUGA ........((((...((((........((((...((((---------(((.......))).))))(((((((.((.((((.....)))).))...)))))))))))...))))...)))) ( -38.00) >DroSim_CAF1 17354 119 - 1 UUUUCCCCUCAUUUCCCGCGCUUUUCCCCGCAUUUCCCCGCAUUUCCCGGCAAAGUCCCGUGGGGCGACCAGUGUCCAGCACUUUGUUGCGCUUUCUGGUCA-UGAUCAGCGGCCCGUGA ........((((...((((......((((((((((..(((.......)))..))))...)))))).((((((.((.((((.....)))).))...)))))).-......))))...)))) ( -37.40) >DroEre_CAF1 3762 102 - 1 ---------CAUUUUCUGCGCUUUUCCCCGCGUUUCCCC---------GGGAAAAUCCCGCGGGGCGACCAGUGUCCAGCACUUUGUUGCGCUUUCUGGUCGGUGGUCAGCGGCCCGUGA ---------.........((((.....((((...(((((---------(((.....)))).))))(((((((.((.((((.....)))).))...)))))))))))..))))........ ( -40.10) >DroYak_CAF1 1294 108 - 1 -------CGCAUUUCCCGCGCUUUUCCUCGCAUUUCCCCCCGU-----GGAAAAAUCCCUGUGGGUGACCAGUGUCCAGCACUUUGUUGCGCUUUCUGGUCGCUGGUCAGCGGCCCGUGA -------(((....(((((((........)).(((((......-----))))).......)))))(((((((((.(((((.(......).))....))).))))))))))))........ ( -38.40) >consensus UUUU___C_CAUUUCCCGCGCUUUUCCCCGCAUUUCCCC________CGGCAAAAUCCCGUGGGGCGACCAGUGUCCAGCACUUUGUUGCGCUUUCUGGUCGGUGGUCAGCGGCCCGUGA ...............((((......((((((............................))))))(((((((.((.((((.....)))).))...))))))).......))))....... (-28.61 = -28.61 + -0.00)

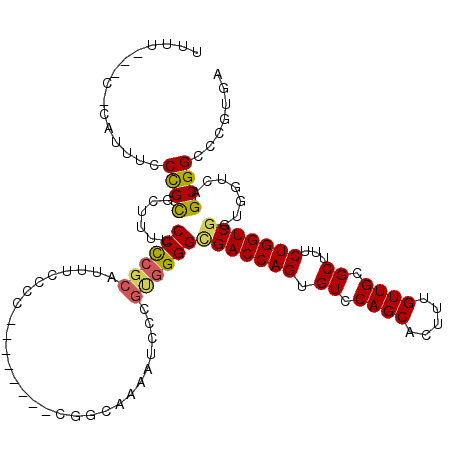

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:10 2006