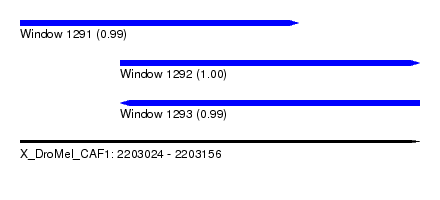
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,203,024 – 2,203,156 |
| Length | 132 |
| Max. P | 0.998936 |

| Location | 2,203,024 – 2,203,116 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.37 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -23.44 |
| Energy contribution | -23.76 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2203024 92 + 22224390 CACAGCUGGAGGUAACGAUGGCCUUUCAACUGGAGAAGUUCUCUGCUCUUGUACGGCAGGAGAUGACUUCUGCAGUGCAGGA---------------------GAAGACGGAU ..(((.(((((((.......))).)))).))).....(((((((.(((((((((.(((((((....))))))).))))))))---------------------).))).)))) ( -36.90) >DroSec_CAF1 1330 92 + 1 CACAGCUGGAGGUAACGAUGGCCUUUCAACUGGAGAAGUCCUUAGCUCUUGUACGGCAGGAGAUGACUUCUGCAGUACAGGA---------------------GAAGACGGAU ..(((.(((((((.......))).)))).))).....((((((..(((((((((.(((((((....))))))).))))))))---------------------)..)).)))) ( -37.00) >DroSim_CAF1 658 92 + 1 CACAGCUGGAGGUAACGAUGGCCUUUCAACUGGAGAAGUUCUUAGCUCUUGUACGGCAGGAGAUGACUUCUGCAGUACAGGA---------------------GAAGACGGAU ..(((.(((((((.......))).)))).))).....((((((..(((((((((.(((((((....))))))).))))))))---------------------)..)).)))) ( -34.10) >DroEre_CAF1 1351 92 + 1 CGCAGCAGGAGGUAACGAUGGCCUUUGAACUGGAGAAGUCCGUUGCUCUUGUACAGCAGGAGAUGACUUCUGCAGCACAGGA---------------------GAGGACGGAU ..(((..((((((.......))))))...))).....(((((((.(((((((.(.(((((((....))))))).).))))))---------------------)..))))))) ( -36.70) >DroYak_CAF1 1405 113 + 1 UACAGCAGGCGGUAACGAUGGCCCUUCAACUGACGAAGAUCGCUGCUCUGGCACUGCAGGCGAUGACUUCUGCAGUACAGGCGAUGACUUCUGCAGUACGGGUGAGGAUGGAU .......(((.((....)).)))(((((((((..((((.(((.(((.(((..((((((((........)))))))).)))))).)))))))..)))).....)))))...... ( -37.70) >consensus CACAGCUGGAGGUAACGAUGGCCUUUCAACUGGAGAAGUUCUUUGCUCUUGUACGGCAGGAGAUGACUUCUGCAGUACAGGA_____________________GAAGACGGAU ...((((((((((.......)))))......(((....))).)))))(((((((.(((((((....))))))).)))))))................................ (-23.44 = -23.76 + 0.32)

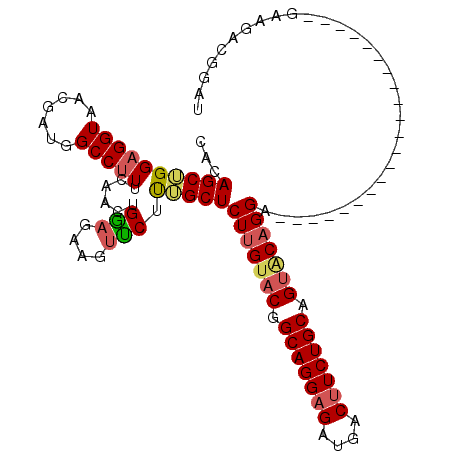
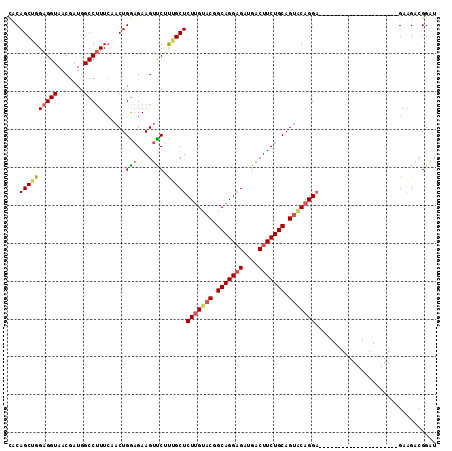
| Location | 2,203,057 – 2,203,156 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.58 |
| Mean single sequence MFE | -39.92 |
| Consensus MFE | -23.62 |
| Energy contribution | -25.02 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.59 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
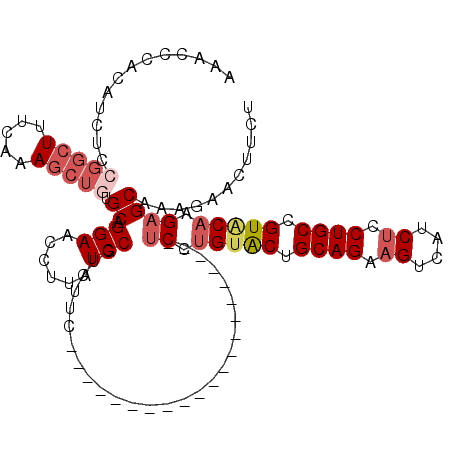
>X_DroMel_CAF1 2203057 99 + 22224390 AGAAGUUCUCUGCUCUUGUACGGCAGGAGAUGACUUCUGCAGUGCAGGA---------------------GAAGACGGAUAAGGUUCCUGCACAGCUCUGAAAGUCGGAGAUGUAGGUUU ....(((((((.(((((((((.(((((((....))))))).))))))))---------------------).))).))))......((((((...(((((.....))))).))))))... ( -42.90) >DroSec_CAF1 1363 99 + 1 AGAAGUCCUUAGCUCUUGUACGGCAGGAGAUGACUUCUGCAGUACAGGA---------------------GAAGACGGAUAAGGUUCCUGCACAGCUUCGAAAGCCGGAGAUGUGGGUUU ....((((((..(((((((((.(((((((....))))))).))))))))---------------------)..)).))))......((..((...(((((.....))))).))..))... ( -40.50) >DroSim_CAF1 691 99 + 1 AGAAGUUCUUAGCUCUUGUACGGCAGGAGAUGACUUCUGCAGUACAGGA---------------------GAAGACGGAUAAGGUUCCUGCACAGCUUCGAAAGCCGGAGAUGUGGGUUU ....((.((...(((((((((.(((((((....))))))).))))))))---------------------).))))(((......)))..((((.(((((.....))))).))))..... ( -38.00) >DroEre_CAF1 1384 99 + 1 AGAAGUCCGUUGCUCUUGUACAGCAGGAGAUGACUUCUGCAGCACAGGA---------------------GAGGACGGAUAAGGUUCCUGCCUAGCUUUGACAGCAGGAGAUGUUGUUUU ....(((((((.(((((((.(.(((((((....))))))).).))))))---------------------)..))))))).((((....))))......(((((((.....))))))).. ( -38.90) >DroYak_CAF1 1438 120 + 1 CGAAGAUCGCUGCUCUGGCACUGCAGGCGAUGACUUCUGCAGUACAGGCGAUGACUUCUGCAGUACGGGUGAGGAUGGAUAGGCUUCCUGCACAGGUUUGACAGCCUGCGAUGUGAUUUG ...((((((((((....))).(((((((...(((((((((((...((.......)).)))))).....(((((((.(......).)))).)))))))).....)))))))..))))))). ( -39.30) >consensus AGAAGUUCUUUGCUCUUGUACGGCAGGAGAUGACUUCUGCAGUACAGGA_____________________GAAGACGGAUAAGGUUCCUGCACAGCUUUGAAAGCCGGAGAUGUGGGUUU ...........((((((((((.(((((((....))))))).))))))))...........................(((......))).))(((.(((((.....))))).)))...... (-23.62 = -25.02 + 1.40)

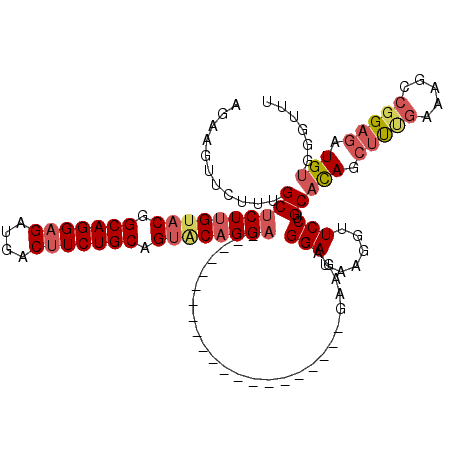
| Location | 2,203,057 – 2,203,156 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.58 |
| Mean single sequence MFE | -29.95 |
| Consensus MFE | -15.84 |
| Energy contribution | -17.36 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2203057 99 - 22224390 AAACCUACAUCUCCGACUUUCAGAGCUGUGCAGGAACCUUAUCCGUCUUC---------------------UCCUGCACUGCAGAAGUCAUCUCCUGCCGUACAAGAGCAGAGAACUUCU ........................((.((((((((...............---------------------)))))))).))((((((..((((.((((......).))))))))))))) ( -28.76) >DroSec_CAF1 1363 99 - 1 AAACCCACAUCUCCGGCUUUCGAAGCUGUGCAGGAACCUUAUCCGUCUUC---------------------UCCUGUACUGCAGAAGUCAUCUCCUGCCGUACAAGAGCUAAGGACUUCU .....((((.((.((.....)).)).))))..(((......)))((((((---------------------((.(((((.((((.((....)).)))).))))).)))...))))).... ( -27.90) >DroSim_CAF1 691 99 - 1 AAACCCACAUCUCCGGCUUUCGAAGCUGUGCAGGAACCUUAUCCGUCUUC---------------------UCCUGUACUGCAGAAGUCAUCUCCUGCCGUACAAGAGCUAAGAACUUCU .....((((.((.((.....)).)).))))..(((......))).(((((---------------------((.(((((.((((.((....)).)))).))))).)))..))))...... ( -25.40) >DroEre_CAF1 1384 99 - 1 AAAACAACAUCUCCUGCUGUCAAAGCUAGGCAGGAACCUUAUCCGUCCUC---------------------UCCUGUGCUGCAGAAGUCAUCUCCUGCUGUACAAGAGCAACGGACUUCU ...........(((((((..........)))))))......(((((...(---------------------((.(((((.((((.((....)).)))).))))).)))..)))))..... ( -33.00) >DroYak_CAF1 1438 120 - 1 CAAAUCACAUCGCAGGCUGUCAAACCUGUGCAGGAAGCCUAUCCAUCCUCACCCGUACUGCAGAAGUCAUCGCCUGUACUGCAGAAGUCAUCGCCUGCAGUGCCAGAGCAGCGAUCUUCG ........(((((...((((...((..(((.((((.(......).)))))))..))...))))........(((((((((((((..(....)..)))))))).))).)).)))))..... ( -34.70) >consensus AAACCCACAUCUCCGGCUUUCAAAGCUGUGCAGGAACCUUAUCCGUCUUC_____________________UCCUGUACUGCAGAAGUCAUCUCCUGCCGUACAAGAGCAAAGAACUUCU .............(((((.....))))).((.(((......)))...........................((.(((((.((((.((....)).)))).))))).))))........... (-15.84 = -17.36 + 1.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:03 2006