
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,849,455 – 19,849,596 |
| Length | 141 |
| Max. P | 0.999941 |

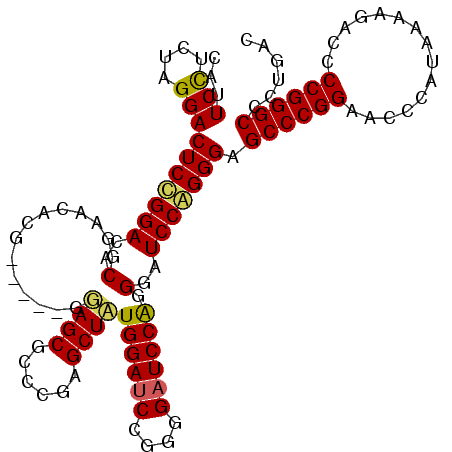
| Location | 19,849,455 – 19,849,567 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 67.67 |
| Mean single sequence MFE | -40.07 |
| Consensus MFE | -27.61 |
| Energy contribution | -28.23 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.69 |
| SVM decision value | 4.71 |
| SVM RNA-class probability | 0.999941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
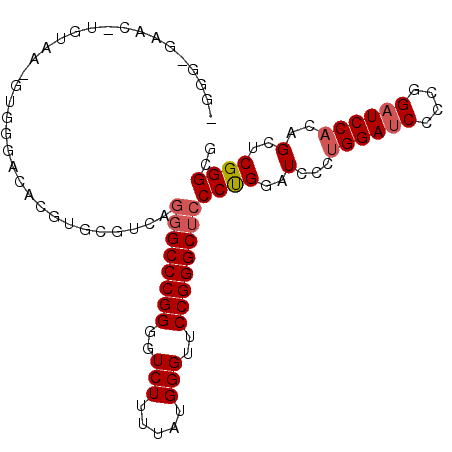
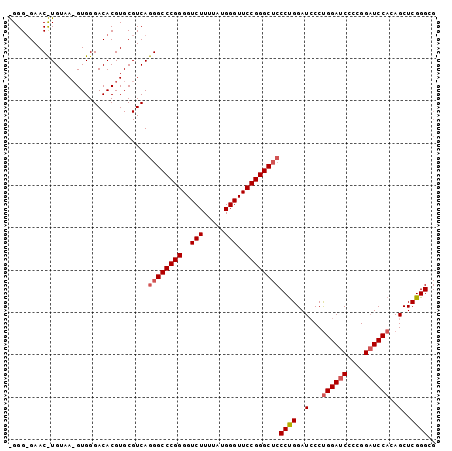
>X_DroMel_CAF1 19849455 112 + 22224390 CAUUCCUCUAGGACUCCGGACACUGUAGACACUUGUGUAGCUCCCGAGCUGUGGAUCCGGGGAUCCGGGGAUCCAGGGAGCCCGGAACCCAUAAAAGACCCCGGGCCCUGAC ...(((((..(((((((((((((.....(((....)))((((....)))))))..))))))).))))))))..((((..((((((...............)))))))))).. ( -44.26) >DroSec_CAF1 7254 107 + 1 CAUUCUUCUAGGACUCUGGACACGAAACACG-----GGAGCGCCCGAGCUAUGGAUCCUGGGAUCCAGGGAUCCAGGGAGCCCGGAACCCAUAAAAGACCCCGGGCCCUGAC ........((((.(((((((..(......((-----((....)))).....((((((....)))))).)..))))))).((((((...............)))))))))).. ( -42.66) >DroSim_CAF1 3957 107 + 1 CAUUCCUCUAGGACUCCGGACACGGAACACG-----GGAGCGCCCGAGCUCUGGAGCCGGGGAUCCAGGCAUCCGGGGANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN ...(((((..(((((((((...((((.(.((-----((....)))).).))))...)))))).))).((...)))))))................................. ( -33.30) >consensus CAUUCCUCUAGGACUCCGGACACGGAACACG_____GGAGCGCCCGAGCUAUGGAUCCGGGGAUCCAGGGAUCCAGGGAGCCCGGAACCCAUAAAAGACCCCGGGCCCUGAC ...(((....)))(((((((..(..............((((......))))((((((....)))))).)..))))))).((((((...............))))))...... (-27.61 = -28.23 + 0.62)

| Location | 19,849,455 – 19,849,567 |
|---|---|
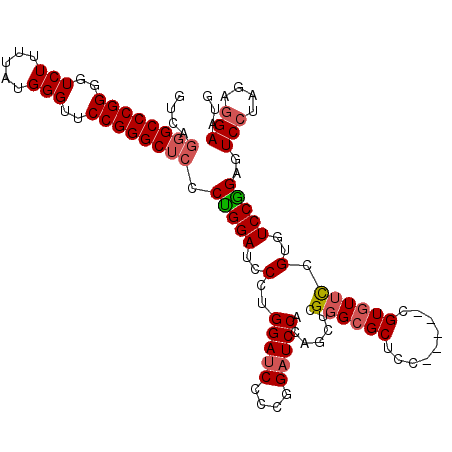
| Length | 112 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 67.67 |
| Mean single sequence MFE | -41.33 |
| Consensus MFE | -29.15 |
| Energy contribution | -31.23 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.997379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19849455 112 - 22224390 GUCAGGGCCCGGGGUCUUUUAUGGGUUCCGGGCUCCCUGGAUCCCCGGAUCCCCGGAUCCACAGCUCGGGAGCUACACAAGUGUCUACAGUGUCCGGAGUCCUAGAGGAAUG ....((((((((..(((.....)))..))))))))((((((((....)))))(((((..(((((((....))))(((....))).....))))))))........))).... ( -47.10) >DroSec_CAF1 7254 107 - 1 GUCAGGGCCCGGGGUCUUUUAUGGGUUCCGGGCUCCCUGGAUCCCUGGAUCCCAGGAUCCAUAGCUCGGGCGCUCC-----CGUGUUUCGUGUCCAGAGUCCUAGAAGAAUG .(((((((.((((((((.....(((.((((((...)))))).))).))))))).(((..(..(((.((((....))-----)).)))..)..))).).))))).))...... ( -46.60) >DroSim_CAF1 3957 107 - 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNUCCCCGGAUGCCUGGAUCCCCGGCUCCAGAGCUCGGGCGCUCC-----CGUGUUCCGUGUCCGGAGUCCUAGAGGAAUG ....................................((((((((.((((.(....).))))((((.((((....))-----)).)))).))))))))..(((....)))... ( -30.30) >consensus GUCAGGGCCCGGGGUCUUUUAUGGGUUCCGGGCUCCCUGGAUCCCUGGAUCCCCGGAUCCACAGCUCGGGCGCUCC_____CGUGUUCCGUGUCCGGAGUCCUAGAGGAAUG ....((((((((..(((.....)))..)))))))).(((((..(..(((((....))))).......((((((.........)))))).)..)))))..(((....)))... (-29.15 = -31.23 + 2.09)


| Location | 19,849,495 – 19,849,596 |
|---|---|
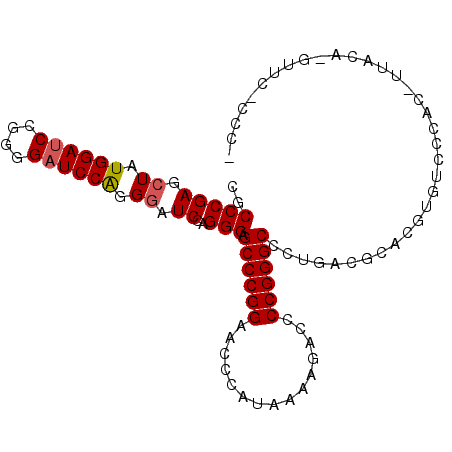
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 52.81 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -17.85 |
| Energy contribution | -18.79 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19849495 101 + 22224390 CUCCCGAGCUGUGGAUCCGGGGAUCCGGGGAUCCAGGGAGCCCGGAACCCAUAAAAGACCCCGGGCCCUGACGCACGUGUCCCACGUUACAAGUUCUCCCU .....(((((((((..((((((.((..(((.(((.((....))))).)))......))))))))..))......(((((...))))).)).)))))..... ( -38.20) >DroSec_CAF1 7289 101 + 1 CGCCCGAGCUAUGGAUCCUGGGAUCCAGGGAUCCAGGGAGCCCGGAACCCAUAAAAGACCCCGGGCCCUGACGCACGUGUCCCACUUUACAGGUUCACCCC .....(((((.((((((....))))))(((((.((((..((((((...............))))))))))((....)))))))........)))))..... ( -37.76) >DroSim_CAF1 3992 101 + 1 CGCCCGAGCUCUGGAGCCGGGGAUCCAGGCAUCCGGGGANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN ..((((.((.(((((.(....).)))))))...))))................................................................ ( -16.80) >consensus CGCCCGAGCUAUGGAUCCGGGGAUCCAGGGAUCCAGGGAGCCCGGAACCCAUAAAAGACCCCGGGCCCUGACGCACGUGUCCCAC_UUACA_GUUC_CCC_ ..(((((.((.((((((....)))))).)).))..))).((((((...............))))))................................... (-17.85 = -18.79 + 0.95)


| Location | 19,849,495 – 19,849,596 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 52.81 |
| Mean single sequence MFE | -35.63 |
| Consensus MFE | -22.47 |
| Energy contribution | -24.33 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19849495 101 - 22224390 AGGGAGAACUUGUAACGUGGGACACGUGCGUCAGGGCCCGGGGUCUUUUAUGGGUUCCGGGCUCCCUGGAUCCCCGGAUCCCCGGAUCCACAGCUCGGGAG (((((...((((..(((((...)))))....))))((((((..(((.....)))..)))))))))))...((((.(((((....))))).......)))). ( -44.60) >DroSec_CAF1 7289 101 - 1 GGGGUGAACCUGUAAAGUGGGACACGUGCGUCAGGGCCCGGGGUCUUUUAUGGGUUCCGGGCUCCCUGGAUCCCUGGAUCCCAGGAUCCAUAGCUCGGGCG ((((((..((........))..))).....(((((((((((..(((.....)))..))))))..)))))..)))((((((....))))))..((....)). ( -45.40) >DroSim_CAF1 3992 101 - 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNUCCCCGGAUGCCUGGAUCCCCGGCUCCAGAGCUCGGGCG ................................................................(((((..(((((((.(....).))))).))))))).. ( -16.90) >consensus _GGG_GAAC_UGUAA_GUGGGACACGUGCGUCAGGGCCCGGGGUCUUUUAUGGGUUCCGGGCUCCCUGGAUCCCUGGAUCCCCGGAUCCACAGCUCGGGCG .................................((((((((..(((.....)))..))))))))((((..(...((((((....))))))..)..)))).. (-22.47 = -24.33 + 1.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:56 2006