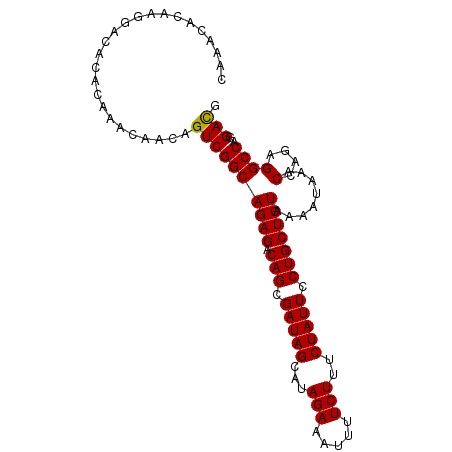
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,847,447 – 19,847,584 |
| Length | 137 |
| Max. P | 0.930646 |

| Location | 19,847,447 – 19,847,553 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 97.48 |
| Mean single sequence MFE | -29.61 |
| Consensus MFE | -29.55 |
| Energy contribution | -29.33 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.92 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19847447 106 + 22224390 CAGCUGGCUGUCCUGCUCCUCCGCACCUUCGUCCUUCGCUCAUCCUGGCCUCUGGUUUAUUUUAGAGCAGGAAUAGAAAGAAAAUUUCUAUGCUAUCGCUGUCUCU ((((((((..((((((((......(((...(.((............)).)...)))........))))))))(((((((.....)))))))))))..))))..... ( -28.84) >DroSec_CAF1 5301 106 + 1 CAGCUGGCUGUCCUGCUCCUCCGCACCUUCGUCCUUUGCCCGUCCUGGCCUCUGGUUUAUUUUAGAGCAGGAAUAGAAAGAAAAUUUCUAUGCUAUCGCUGUCUCU ((((((((..((((((((....((......))((...(((......)))....)).........))))))))(((((((.....)))))))))))..))))..... ( -30.00) >DroSim_CAF1 1973 106 + 1 CAGCUGGCUGUCCUGCUCCGCCGCACCUUCGUCCUUUGCCCGUCCUGGCCUCUGGUUUAUUUUAGAGCAGGAAUAGAAAGAAAAUUUCUAUGCUAUCGCUGUCUCU ((((((((..((((((((....((......))((...(((......)))....)).........))))))))(((((((.....)))))))))))..))))..... ( -30.00) >consensus CAGCUGGCUGUCCUGCUCCUCCGCACCUUCGUCCUUUGCCCGUCCUGGCCUCUGGUUUAUUUUAGAGCAGGAAUAGAAAGAAAAUUUCUAUGCUAUCGCUGUCUCU ((((((((..((((((((....((......))((...(((......)))....)).........))))))))(((((((.....)))))))))))..))))..... (-29.55 = -29.33 + -0.22)

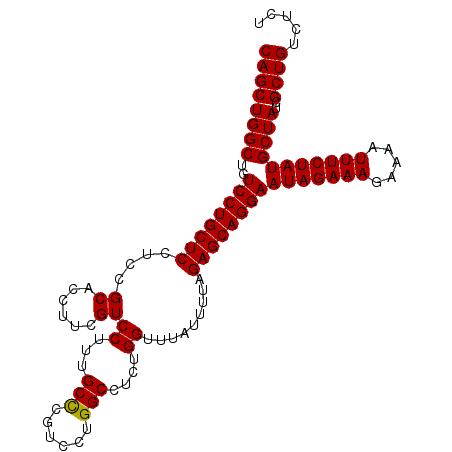

| Location | 19,847,447 – 19,847,553 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 97.48 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -32.30 |
| Energy contribution | -32.63 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19847447 106 - 22224390 AGAGACAGCGAUAGCAUAGAAAUUUUCUUUCUAUUCCUGCUCUAAAAUAAACCAGAGGCCAGGAUGAGCGAAGGACGAAGGUGCGGAGGAGCAGGACAGCCAGCUG .....((((....(((((((((.....)))))))(((((((((........((........)).........(.((....)).)...)))))))))..))..)))) ( -30.60) >DroSec_CAF1 5301 106 - 1 AGAGACAGCGAUAGCAUAGAAAUUUUCUUUCUAUUCCUGCUCUAAAAUAAACCAGAGGCCAGGACGGGCAAAGGACGAAGGUGCGGAGGAGCAGGACAGCCAGCUG .....((((....(((((((((.....)))))))(((((((((..............(((......)))...(.((....)).)...)))))))))..))..)))) ( -34.20) >DroSim_CAF1 1973 106 - 1 AGAGACAGCGAUAGCAUAGAAAUUUUCUUUCUAUUCCUGCUCUAAAAUAAACCAGAGGCCAGGACGGGCAAAGGACGAAGGUGCGGCGGAGCAGGACAGCCAGCUG .....((((....(((((((((.....)))))))(((((((((..............(((......)))...(.((....)).)...)))))))))..))..)))) ( -34.20) >consensus AGAGACAGCGAUAGCAUAGAAAUUUUCUUUCUAUUCCUGCUCUAAAAUAAACCAGAGGCCAGGACGGGCAAAGGACGAAGGUGCGGAGGAGCAGGACAGCCAGCUG .....((((....(((((((((.....)))))))(((((((((..............(((......)))...(.((....)).)...)))))))))..))..)))) (-32.30 = -32.63 + 0.33)

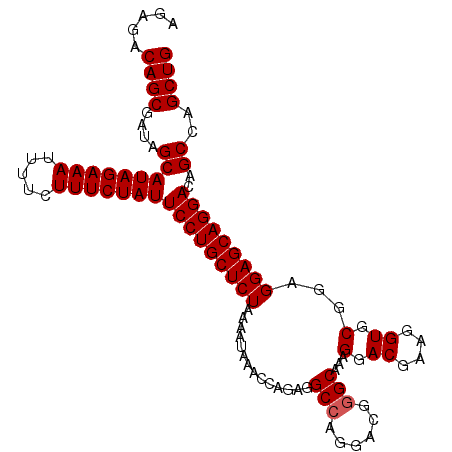

| Location | 19,847,487 – 19,847,584 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 88.32 |
| Mean single sequence MFE | -17.00 |
| Consensus MFE | -16.69 |
| Energy contribution | -16.47 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19847487 97 - 22224390 CAAACACAAGGACACACAAACAACAGUCGGCAGAGACAGCGAUAGCAUAGAAAUUUUCUUUCUAUUCCUGCUCUAAAAUAAACCAGAGGCCAGGAUG ..........(((............)))(((((((.(((.(((((...(((.....)))..))))).)))))))........(....))))...... ( -16.60) >DroSec_CAF1 5341 81 - 1 C----------------AAACAACAGUCGGCAGAGACAGCGAUAGCAUAGAAAUUUUCUUUCUAUUCCUGCUCUAAAAUAAACCAGAGGCCAGGACG .----------------........((((((((((.(((.(((((...(((.....)))..))))).)))))))........(....))))..))). ( -17.20) >DroSim_CAF1 2013 97 - 1 CAAACACAAGGACACACAAACAACAGUCGGCAGAGACAGCGAUAGCAUAGAAAUUUUCUUUCUAUUCCUGCUCUAAAAUAAACCAGAGGCCAGGACG .........................((((((((((.(((.(((((...(((.....)))..))))).)))))))........(....))))..))). ( -17.20) >consensus CAAACACAAGGACACACAAACAACAGUCGGCAGAGACAGCGAUAGCAUAGAAAUUUUCUUUCUAUUCCUGCUCUAAAAUAAACCAGAGGCCAGGACG .........................((((((((((.(((.(((((...(((.....)))..))))).)))))))........(....))))..))). (-16.69 = -16.47 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:53 2006