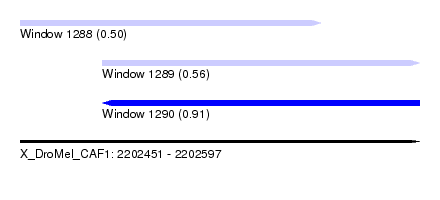
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,202,451 – 2,202,597 |
| Length | 146 |
| Max. P | 0.908982 |

| Location | 2,202,451 – 2,202,561 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.61 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -21.20 |
| Energy contribution | -20.53 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.54 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
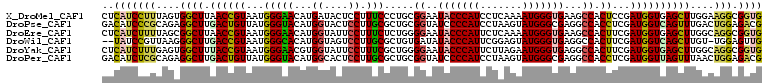
>X_DroMel_CAF1 2202451 110 + 22224390 CUCAUCCUUUAGUGGCUUAACCGUAAUGGGAACAUGAUACUCCUUUCCCUGCGGAAUACCCAUCCUCAAAAUGGGUGAAGCCACUCCGAUGGUGAGCUUGGAAGGCGGUG .(((((.((.((((((((..(((((..(((((............))))))))))..(((((((.......)))))))))))))))..)).)))))(((.....))).... ( -37.60) >DroPse_CAF1 892 110 + 1 GACAUCCCGCAGAGGCUUGACUGUUAUGGGUACAUGGUACUCCUUGCGCUGCGGUAUCCCCAUCCUAAGUAUGGGCGAGGCCACCUCGAUGGUCAGUUUGACUGGAGACG ..((((((((((..((..((.((..(((....)))..)).))...)).))))))....(((((.......))))).((((...))))))))(((.....))).(....). ( -33.80) >DroEre_CAF1 781 110 + 1 CUCAUCUUUUAGCGGCUUAACCGUAAUGGGAACAUGGUAUUCCUUUCUCUGGGGAAUACCCAUUCUCAAAAUGGGUGAAGCCACUUCGAUGGUGAGCUUGGCAGGCGGUG ...........((.((((..((((..((((((...((((((((((.....))))))))))..))))))..))))((.((((((((.....)))).)))).)))))).)). ( -41.90) >DroWil_CAF1 2471 107 + 1 --UAUCCGUUAAGGGCUUGACCGUAAUGGGCACAUGGUAGUCCUUGCGCUGUGAUAUACCCAUUCGGAGUAUGGGUGAGGCCACUUCGAUGGUCAGCUUGU-UGGAGUUG --..((((....(((((.((((((....(((.((((((.((....))))))))...(((((((.......)))))))..)))......)))))))))))..-)))).... ( -36.40) >DroYak_CAF1 799 110 + 1 CUCAUCUUUGAGUGGCUUUACCGUAAUGGGAACGUGGUAUUCCUUUCGCUGGGGAAUACCCAUUCUUAGAAUGGGUGAGGCCACUUCGAUGGUGAGCUUGGCAGGCGGUG .(((((.(((((((((((((((((..((((((.(.((((((((((.....))))))))))).))))))..)).))))))))))).)))).)))))(((.....))).... ( -49.10) >DroPer_CAF1 892 110 + 1 GACAUCUCGCAGAGGCUUGACUGUUAUGGGUACAUGGCACUCCUUGCGCUGCGGUAUCCCCAUCCUAAGUAUGGGCGAGGCCACCUCGAUGGUUAGUUUAACUGGAGACG ..((((..((((..((..((.(((((((....))))))).))...)).))))(((.(((((((.......))))).)).))).....))))............(....). ( -35.10) >consensus CUCAUCCUUUAGAGGCUUGACCGUAAUGGGAACAUGGUACUCCUUGCGCUGCGGAAUACCCAUCCUAAGAAUGGGUGAGGCCACUUCGAUGGUGAGCUUGGCAGGAGGUG ..(((((((....((((.((((((...(((((...((....)).))).....((..(((((((.......)))))))...)).))...))))))))))....))).)))) (-21.20 = -20.53 + -0.66)
| Location | 2,202,481 – 2,202,597 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.95 |
| Mean single sequence MFE | -43.30 |
| Consensus MFE | -14.18 |
| Energy contribution | -14.13 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
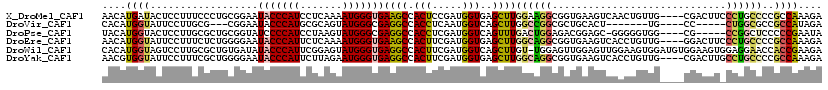
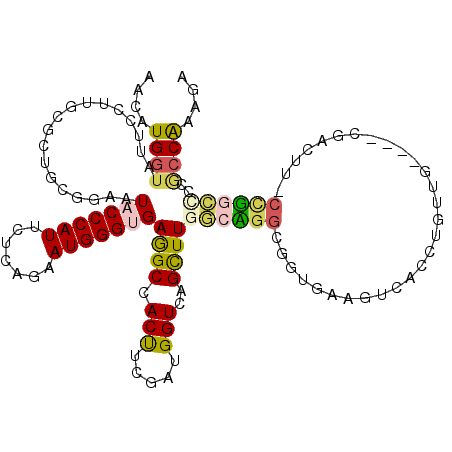
>X_DroMel_CAF1 2202481 116 + 22224390 AACAUGAUACUCCUUUCCCUGCGGAAUACCCAUCCUCAAAAUGGGUGAAGCCACUCCGAUGGUGAGCUUGGAAGGCGGUGAAGUCAACUGUUG----CGACUUCCCUGCCCCGCCAAAGA ............((((....((((..(((((((.......)))))))((((((((.....)))).))))....(((((.((((((........----.)))))).))))))))).)))). ( -37.80) >DroVir_CAF1 886 101 + 1 CACAUGGUAUUCCUUGCG---CGGAAUACCCAUGCGCAGUAUGGGCGAGGCCACCUCAAUGGUCAGCUUGGCCGGCGCUGCACU-------UG----CC-----CUGGCGCCGCCAUAGA .....((((((((.....---.)))))))).(((.((......(((.((((.(((.....)))..)))).)))((((((((...-------.)----).-----..)))))))))))... ( -42.20) >DroPse_CAF1 922 110 + 1 UACAUGGUACUCCUUGCGCUGCGGUAUCCCCAUCCUAAGUAUGGGCGAGGCCACCUCGAUGGUCAGUUUGACUGGAGACGGAGC-GGGGGUGG----CG-----CCGGCUCCCCCGAAUA ..((.((....)).))((((..(((.(((((((.......))))).)).))).((((..(((((.....)))))..)).)))))-)((((..(----(.-----...))..))))..... ( -39.40) >DroEre_CAF1 811 116 + 1 AACAUGGUAUUCCUUUCUCUGGGGAAUACCCAUUCUCAAAAUGGGUGAAGCCACUUCGAUGGUGAGCUUGGCAGGCGGUGAAGUCACCUGUUG----GGACUUCCCUGCCCCGCCAAAGA .....((((((((((.....))))))))))(((((.......)))))((((((((.....)))).))))(((.(((((.((((((.((....)----))))))).)))))..)))..... ( -49.80) >DroWil_CAF1 2499 119 + 1 CACAUGGUAGUCCUUGCGCUGUGAUAUACCCAUUCGGAGUAUGGGUGAGGCCACUUCGAUGGUCAGCUUGU-UGGAGUUGGAGUUGGAAGUGGAUGUGGAAGUGGAGGAACCACCGAAGA ....((((..(((((.((((.(.((((((((((.......))))))....(((((((.(...(((((((..-..)))))))...).))))))))))).).)))))))))...)))).... ( -41.70) >DroYak_CAF1 829 116 + 1 AACGUGGUAUUCCUUUCGCUGGGGAAUACCCAUUCUUAGAAUGGGUGAGGCCACUUCGAUGGUGAGCUUGGCAGGCGGUGAAGUCACCUGUUG----CGACUUGCCUGCCCCGCCAAAGA ...(((((.((((((.....))))))(((((((((...)))))))))..))))).((..(((((.(...(((((((((((....)))).((..----..))..)))))))))))))..)) ( -48.90) >consensus AACAUGGUAUUCCUUGCGCUGCGGAAUACCCAUUCUCAGAAUGGGUGAGGCCACUUCGAUGGUCAGCUUGGCAGGCGGUGAAGUCACCUGUUG____CGACUU_CCGGCCCCGCCAAAGA ....((((..................(((((((.......)))))))((((.(((.....)))..))))((((((.............................))))))..)))).... (-14.18 = -14.13 + -0.05)
| Location | 2,202,481 – 2,202,597 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.95 |
| Mean single sequence MFE | -40.80 |
| Consensus MFE | -16.56 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
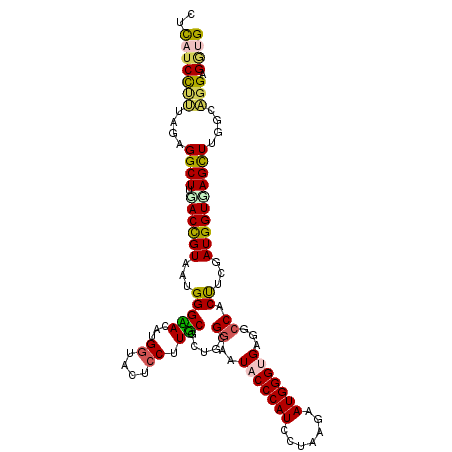
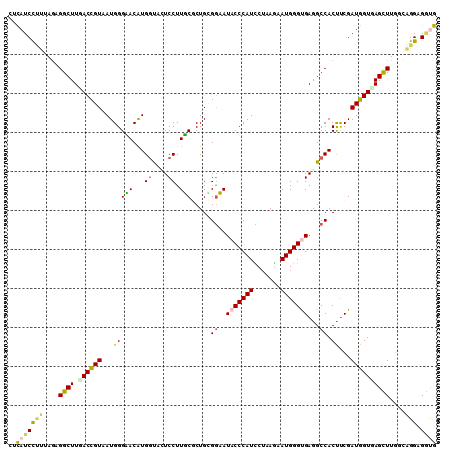
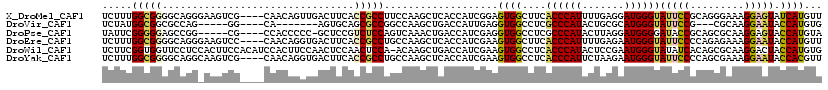
>X_DroMel_CAF1 2202481 116 - 22224390 UCUUUGGCGGGGCAGGGAAGUCG----CAACAGUUGACUUCACCGCCUUCCAAGCUCACCAUCGGAGUGGCUUCACCCAUUUUGAGGAUGGGUAUUCCGCAGGGAAAGGAGUAUCAUGUU ..(((((.(((((.(((((((((----(....).))))))).))))))))))))..((((.(((((((((......))))))))))).))((((((((.........))))))))..... ( -42.60) >DroVir_CAF1 886 101 - 1 UCUAUGGCGGCGCCAG-----GG----CA-------AGUGCAGCGCCGGCCAAGCUGACCAUUGAGGUGGCCUCGCCCAUACUGCGCAUGGGUAUUCCG---CGCAAGGAAUACCAUGUG .....((((((((...-----.(----(.-------...)).)))))(((((..((........)).)))))..))).......(((((((.((((((.---.....))))))))))))) ( -42.90) >DroPse_CAF1 922 110 - 1 UAUUCGGGGGAGCCGG-----CG----CCACCCCC-GCUCCGUCUCCAGUCAAACUGACCAUCGAGGUGGCCUCGCCCAUACUUAGGAUGGGGAUACCGCAGCGCAAGGAGUACCAUGUA .....(((((.((...-----.)----)..)))))-(((((.....(((.....))).....((..((((..((.(((((.(....))))))))..))))..))...)))))........ ( -37.50) >DroEre_CAF1 811 116 - 1 UCUUUGGCGGGGCAGGGAAGUCC----CAACAGGUGACUUCACCGCCUGCCAAGCUCACCAUCGAAGUGGCUUCACCCAUUUUGAGAAUGGGUAUUCCCCAGAGAAAGGAAUACCAUGUU (((((((.(((((((((((((((----(....)).))))))....))))))..(((((...(((((((((......)))))))))...)))))...)))))))))..((....))..... ( -48.50) >DroWil_CAF1 2499 119 - 1 UCUUCGGUGGUUCCUCCACUUCCACAUCCACUUCCAACUCCAACUCCA-ACAAGCUGACCAUCGAAGUGGCCUCACCCAUACUCCGAAUGGGUAUAUCACAGCGCAAGGACUACCAUGUG .....((((((.(((............(((((((......((.((...-...)).))......)))))))....((((((.......)))))).............)))))))))..... ( -27.90) >DroYak_CAF1 829 116 - 1 UCUUUGGCGGGGCAGGCAAGUCG----CAACAGGUGACUUCACCGCCUGCCAAGCUCACCAUCGAAGUGGCCUCACCCAUUCUAAGAAUGGGUAUUCCCCAGCGAAAGGAAUACCACGUU .....(((..(((((((((((((----(.....)))))))....)))))))..)))..........((((....((((((((...))))))))(((((.........))))).))))... ( -45.40) >consensus UCUUUGGCGGGGCAGG_AAGUCG____CAACAGCUGACUUCACCGCCUGCCAAGCUCACCAUCGAAGUGGCCUCACCCAUACUGAGAAUGGGUAUUCCGCAGCGAAAGGAAUACCAUGUU .....(((((................................)))))...................((((....((((((.......))))))(((((.........))))).))))... (-16.56 = -16.73 + 0.17)
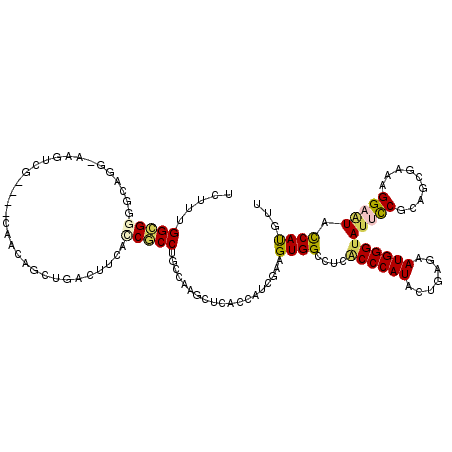
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:00 2006