| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,841,634 – 19,841,735 |
| Length | 101 |
| Max. P | 0.537199 |

| Location | 19,841,634 – 19,841,735 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.23 |
| Mean single sequence MFE | -30.94 |
| Consensus MFE | -22.37 |
| Energy contribution | -22.53 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19841634 101 + 22224390 GUUGCGUGCCAAAGUCAUGGCCUCCUGCACUCCGUACCGCACAAAGGACAAUGGCUUCAUGGCGACGCUGCGA-ACGGAA--GUCGGUCGGAUUCGGAAUG-AGA ((.(((((((((((((((..(((..(((..........)))...)))...)))))))..)))).)))).))..-.(.(((--..(....)..))).)....-... ( -28.80) >DroVir_CAF1 16372 99 + 1 GUUGCGUGCCAAAGUCAUGGCCUCCUGGACGCCGUAGCGCACAAAGGACAAUGGCUUCAUGGCAACGCUGCUG-CUGCAAAACACAAGAGCAA-AGCA----CAC ...(((((((((((((((..(((..((..(((....)))..)).)))...)))))))..)))).))))((((.-.(((...........))).-))))----... ( -34.20) >DroPse_CAF1 748 101 + 1 GUUGCGUGCCAAAGUCAUGGCCUCCUGCACUCCAUAGCGGACAAAGGACAAUGGCUUCAUGGCGACGCUGCAAAACGGAG--CACAAACGGAA-CGGAACC-GCA (((.((((((((((((((..(((.((((........))))....)))...)))))))..))))...(((.(.....).))--)....))).))-)......-... ( -31.00) >DroGri_CAF1 65676 102 + 1 GUUGCGUGCCAAAGUCAUGGCCUCCUGCACUCCGUAGCGUACAAAGGACAAUGGCUUCAUGGCGACGCUGCUG-CUAAAAAACACAAGAGCAA-AGGAGCA-AAC ...(((((((((((((((..(((..(((.((....)).)))...)))...)))))))..)))).))))((((.-((.................-)).))))-... ( -28.53) >DroMoj_CAF1 22518 102 + 1 GUUGCGUGCCAAAGUCAUGGCCUCCUGCACACCGUAGCGCACAAAGGACAAUGGCUUCAUGGCGACACUGCUG-CUGUAAGGCACAAGAGCAG-AAAA-CAACAA ((((.(((((((((((((..(((..(((.(......).)))...)))...)))))))..)))).)).(((((.-.(((.....)))..)))))-....-)))).. ( -32.10) >DroPer_CAF1 754 100 + 1 GUUGCGUGCCAAAGUCAUGGCCUCCUGCACUCCAUAGCGGACAAAGGACAAUGGCUUCAUGGCGACGCUGCAA-ACGGAG--CACAAACGGAA-CGGANNN-NNN (((.((((((((((((((..(((.((((........))))....)))...)))))))..))))...(((.(..-..).))--)....))).))-)......-... ( -31.00) >consensus GUUGCGUGCCAAAGUCAUGGCCUCCUGCACUCCGUAGCGCACAAAGGACAAUGGCUUCAUGGCGACGCUGCUA_ACGGAA__CACAAGAGCAA_AGGA_C__AAA ((.(((((((((((((((..(((..((..(........)..)).)))...)))))))..)))).)))).)).................................. (-22.37 = -22.53 + 0.17)

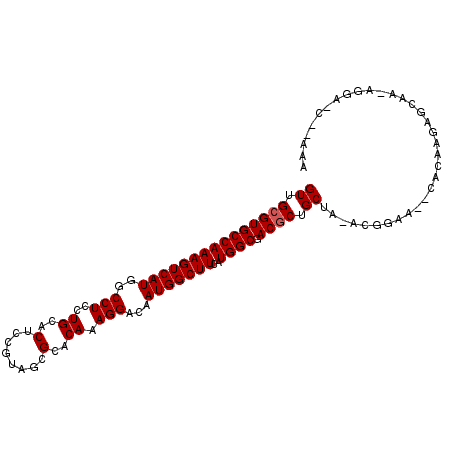
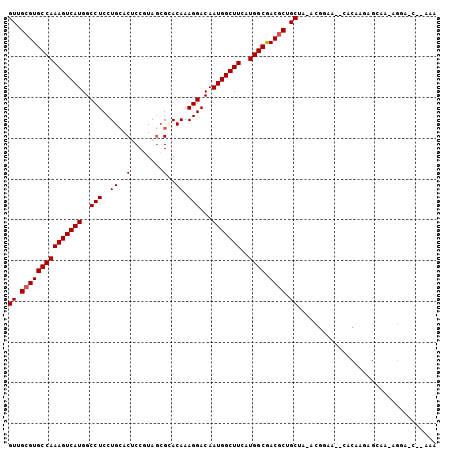
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:47 2006