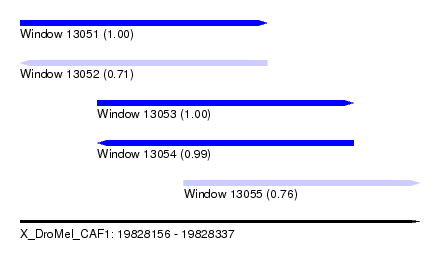
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,828,156 – 19,828,337 |
| Length | 181 |
| Max. P | 0.999813 |

| Location | 19,828,156 – 19,828,268 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 88.49 |
| Mean single sequence MFE | -46.20 |
| Consensus MFE | -37.10 |
| Energy contribution | -38.62 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19828156 112 + 22224390 UUCACAAUCGCGGGAUAGGGCUACUCCGGGUUCUCCGUGUACUCUGGGUU-UUCCGGUCUUUCCGGGUAUUCCGGCGCCUGUCAGCUGGCAGUUUUCCUUUACCAGCUGGCAG ........(((.((((((((.(((..(((.....))).))))))))....-.(((((.....)))))...))).))).(((((((((((.((.......)).))))))))))) ( -42.90) >DroSec_CAF1 33714 110 + 1 UUCACAAUC--CGGAUAGGGCUACUACGGGUACUCCGGGUACUCUGGGUU-UUCCGCGCUUUCCGGGUAUUCCGGCGCCUGUCAGCUGGCAGUUUUCCUUUGCCAGCUGGCAG ......(((--((((.((.((......((((((.....))))))..((..-..)))).)).)))))))..........((((((((((((((.......)))))))))))))) ( -50.20) >DroSim_CAF1 32065 110 + 1 UUCACAAUC--GGGAUAGGGCUACUACGGGUACUCCGCGUACUCUGGGUU-UUCCGGGCUUUCCGGGUAUUCCGGCGCCUGUCAGCUGGCAGUUUUCCUUUGCCAGCUGGCAG .......((--((((((......(((.((((((.....)))))))))...-.(((((.....)))))))))))))...((((((((((((((.......)))))))))))))) ( -48.20) >DroEre_CAF1 23945 109 + 1 UUCACAAUC--GGGAUAGGGGUACUCCGGGUUUUCCGGGUACUCUGGGUU-UUCCGGGCUUUCCGGGUAUUCCGGCGCCUGUCAUCUGGCAGUUU-CCCCCACCAGCUGGCAG .......((--(((((((((((((.((((.....))))))))))).....-.(((((.....)))))))))))))...((((((.((((..(...-...)..)))).)))))) ( -42.60) >DroYak_CAF1 24828 104 + 1 UUCACAAUC--GGAAUAUAG-------GGGUAUGCCGGGUACUCUGGGUUUUUCCGGGCUUUCCGGCUAUUCCGGCGCCUGUCAGCUGGCUGUUUUCCUUUGCCAGCUGGCAG .........--........(-------(((((.((((((...((((((....))))))...)))))))))))).....((((((((((((...........)))))))))))) ( -47.10) >consensus UUCACAAUC__GGGAUAGGGCUACUACGGGUACUCCGGGUACUCUGGGUU_UUCCGGGCUUUCCGGGUAUUCCGGCGCCUGUCAGCUGGCAGUUUUCCUUUGCCAGCUGGCAG ..........................((((...((((((...((((((....))))))...))))))...))))....((((((((((((((.......)))))))))))))) (-37.10 = -38.62 + 1.52)


| Location | 19,828,156 – 19,828,268 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 88.49 |
| Mean single sequence MFE | -37.61 |
| Consensus MFE | -27.88 |
| Energy contribution | -28.32 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19828156 112 - 22224390 CUGCCAGCUGGUAAAGGAAAACUGCCAGCUGACAGGCGCCGGAAUACCCGGAAAGACCGGAA-AACCCAGAGUACACGGAGAACCCGGAGUAGCCCUAUCCCGCGAUUGUGAA (((.(((((((((.........))))))))).))).(((.(((....((((.....))))..-.....((.((((.(((.....)))..)).)).)).))).)))........ ( -37.10) >DroSec_CAF1 33714 110 - 1 CUGCCAGCUGGCAAAGGAAAACUGCCAGCUGACAGGCGCCGGAAUACCCGGAAAGCGCGGAA-AACCCAGAGUACCCGGAGUACCCGUAGUAGCCCUAUCCG--GAUUGUGAA (((((((((((((.........)))))))))....((.((((.....))))...))))))..-....((.(((..(((((.....(......).....))))--)))).)).. ( -38.80) >DroSim_CAF1 32065 110 - 1 CUGCCAGCUGGCAAAGGAAAACUGCCAGCUGACAGGCGCCGGAAUACCCGGAAAGCCCGGAA-AACCCAGAGUACGCGGAGUACCCGUAGUAGCCCUAUCCC--GAUUGUGAA ....(((((((((.........)))))))))((((((.((((.....))))...)))(((..-.......(.((((.((....)))))).).........))--)..)))... ( -37.97) >DroEre_CAF1 23945 109 - 1 CUGCCAGCUGGUGGGGG-AAACUGCCAGAUGACAGGCGCCGGAAUACCCGGAAAGCCCGGAA-AACCCAGAGUACCCGGAAAACCCGGAGUACCCCUAUCCC--GAUUGUGAA (..((....))..)(((-(..(((........(.(((.((((.....))))...))).)...-....))).((((((((.....)))).)))).....))))--......... ( -37.16) >DroYak_CAF1 24828 104 - 1 CUGCCAGCUGGCAAAGGAAAACAGCCAGCUGACAGGCGCCGGAAUAGCCGGAAAGCCCGGAAAAACCCAGAGUACCCGGCAUACCC-------CUAUAUUCC--GAUUGUGAA (((.((((((((...........)))))))).))).(((((((((((((((...(((.((......)).).))..)))))......-------...))))))--)...))).. ( -37.00) >consensus CUGCCAGCUGGCAAAGGAAAACUGCCAGCUGACAGGCGCCGGAAUACCCGGAAAGCCCGGAA_AACCCAGAGUACCCGGAGUACCCGGAGUAGCCCUAUCCC__GAUUGUGAA (((.(((((((((.........))))))))).)))((.((((.....))))...))..((......))........(((.....))).......................... (-27.88 = -28.32 + 0.44)

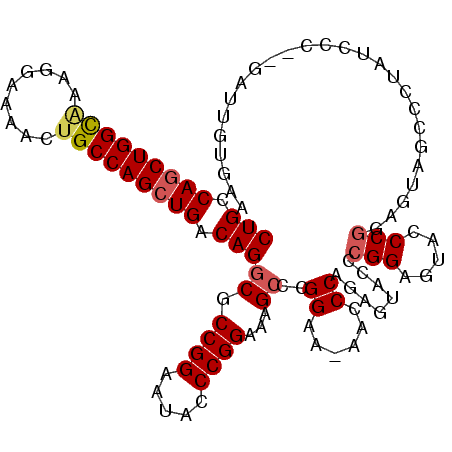
| Location | 19,828,191 – 19,828,307 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 92.96 |
| Mean single sequence MFE | -52.02 |
| Consensus MFE | -45.67 |
| Energy contribution | -47.15 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.14 |
| SVM RNA-class probability | 0.999813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19828191 116 + 22224390 CGUGUACUCUGGGUU-UUCCGGUCUUUCCGGGUAUUCCGGCGCCUGUCAGCUGGCAGUUUUCCUUUACCAGCUGGCAGUGCC-UGGCUGACGUGAAUUCCCGAAAAGUCACAGGGCCU ......(((((((..-..)).(.((((.((((.((((((((((.((((((((((.((.......)).)))))))))))))))-.(.....)).)))).)))).)))).).)))))... ( -46.00) >DroSec_CAF1 33747 113 + 1 CGGGUACUCUGGGUU-UUCCGCGCUUUCCGGGUAUUCCGGCGCCUGUCAGCUGGCAGUUUUCCUUUGCCAGCUGGCAG---C-CGGCUGACGGGAAUUCCCGAAAAGUCACAGGGCCU .(((..(((((((..-..))..(((((.((((.(((((.((((((((((((((((((.......))))))))))))).---.-.))).).).))))).)))).)))))..)))))))) ( -54.20) >DroSim_CAF1 32098 113 + 1 CGCGUACUCUGGGUU-UUCCGGGCUUUCCGGGUAUUCCGGCGCCUGUCAGCUGGCAGUUUUCCUUUGCCAGCUGGCAG---C-CGGCUGACGGGAAUUCCCGAAAAGUCACAGGGCCU ......((((((..(-((.((((..((((.(.....(((((...(((((((((((((.......))))))))))))))---)-)))....).))))..)))).)))..).)))))... ( -56.60) >DroEre_CAF1 23978 112 + 1 CGGGUACUCUGGGUU-UUCCGGGCUUUCCGGGUAUUCCGGCGCCUGUCAUCUGGCAGUUU-CCCCCACCAGCUGGCAG---C-CGGCUGACGGGAGUUCCCGAAAAGUCACAGGGCCU .(((..(((((((..-..)).((((((.((((.(((((((...(((((....)))))...-)).....(((((((...---)-))))))...))))).)))).)))))).)))))))) ( -46.40) >DroYak_CAF1 24854 115 + 1 CGGGUACUCUGGGUUUUUCCGGGCUUUCCGGCUAUUCCGGCGCCUGUCAGCUGGCUGUUUUCCUUUGCCAGCUGGCAG---CCCGGCUGACGGGAAUUCCCGAAAAGUCACAGGGCCU .(((..((((((..(((((.((..((((((......((((.((.(((((((((((...........))))))))))))---)))))....))))))..)).)))))..).)))))))) ( -56.90) >consensus CGGGUACUCUGGGUU_UUCCGGGCUUUCCGGGUAUUCCGGCGCCUGUCAGCUGGCAGUUUUCCUUUGCCAGCUGGCAG___C_CGGCUGACGGGAAUUCCCGAAAAGUCACAGGGCCU ......(((((((.....)).((((((.((((.(((((.((((((((((((((((((.......))))))))))))).......))).).).))))).)))).)))))).)))))... (-45.67 = -47.15 + 1.48)

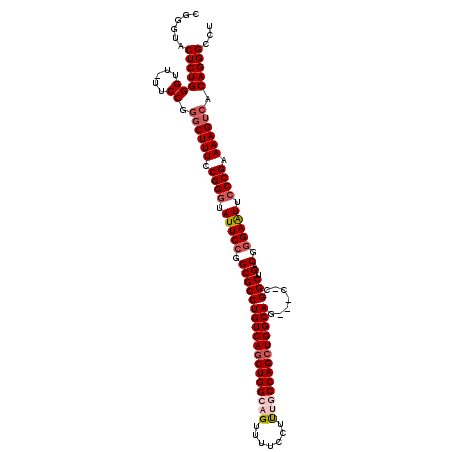
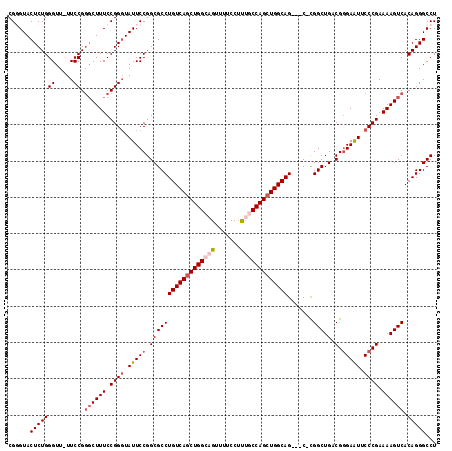
| Location | 19,828,191 – 19,828,307 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 92.96 |
| Mean single sequence MFE | -47.56 |
| Consensus MFE | -38.46 |
| Energy contribution | -38.90 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19828191 116 - 22224390 AGGCCCUGUGACUUUUCGGGAAUUCACGUCAGCCA-GGCACUGCCAGCUGGUAAAGGAAAACUGCCAGCUGACAGGCGCCGGAAUACCCGGAAAGACCGGAA-AACCCAGAGUACACG .......(((.(((((((((.((((..(....)..-(((.(((.(((((((((.........))))))))).)))..))).)))).)))))))))...((..-...))......))). ( -44.10) >DroSec_CAF1 33747 113 - 1 AGGCCCUGUGACUUUUCGGGAAUUCCCGUCAGCCG-G---CUGCCAGCUGGCAAAGGAAAACUGCCAGCUGACAGGCGCCGGAAUACCCGGAAAGCGCGGAA-AACCCAGAGUACCCG .((..(((((.(((((((((.(((((.(.....((-.---(((.(((((((((.........))))))))).))).))).))))).))))))))))))((..-...))..))..)).. ( -49.70) >DroSim_CAF1 32098 113 - 1 AGGCCCUGUGACUUUUCGGGAAUUCCCGUCAGCCG-G---CUGCCAGCUGGCAAAGGAAAACUGCCAGCUGACAGGCGCCGGAAUACCCGGAAAGCCCGGAA-AACCCAGAGUACGCG .......((((((((..(((..((((.((((((.(-(---(((((....)))).((.....))))).)))))).(((.((((.....))))...))).))))-..)))))))).))). ( -48.70) >DroEre_CAF1 23978 112 - 1 AGGCCCUGUGACUUUUCGGGAACUCCCGUCAGCCG-G---CUGCCAGCUGGUGGGGG-AAACUGCCAGAUGACAGGCGCCGGAAUACCCGGAAAGCCCGGAA-AACCCAGAGUACCCG .((..((.((..((((((((..(((((..((((.(-(---...)).))))..)))))-.....(((........))).((((.....))))....)))))))-)...)).))..)).. ( -45.30) >DroYak_CAF1 24854 115 - 1 AGGCCCUGUGACUUUUCGGGAAUUCCCGUCAGCCGGG---CUGCCAGCUGGCAAAGGAAAACAGCCAGCUGACAGGCGCCGGAAUAGCCGGAAAGCCCGGAAAAACCCAGAGUACCCG .((..((.((..((((((((..((((.((...(((((---((..((((((((...........))))))))...))).))))....)).))))..))))))))....)).))..)).. ( -50.00) >consensus AGGCCCUGUGACUUUUCGGGAAUUCCCGUCAGCCG_G___CUGCCAGCUGGCAAAGGAAAACUGCCAGCUGACAGGCGCCGGAAUACCCGGAAAGCCCGGAA_AACCCAGAGUACCCG .(((((((((.((...((((....))))..)).)..........(((((((((.........)))))))))))))).)))((.....((((.....))))......)).......... (-38.46 = -38.90 + 0.44)

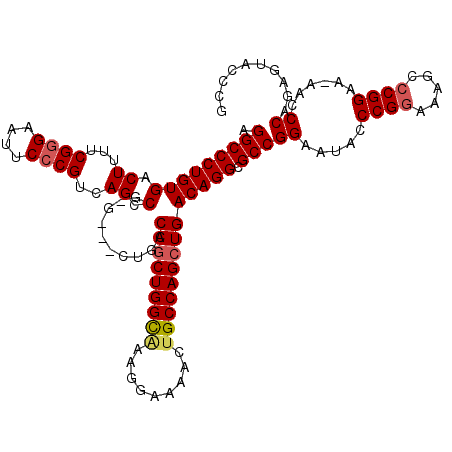
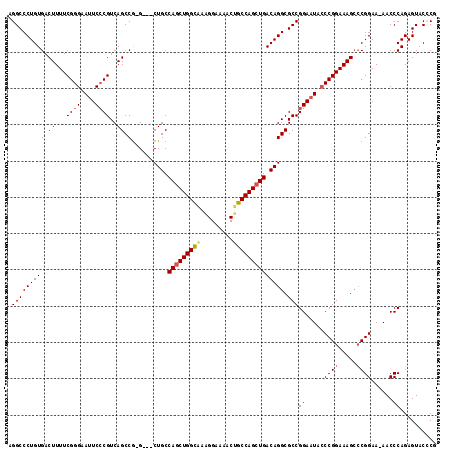
| Location | 19,828,230 – 19,828,337 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.88 |
| Mean single sequence MFE | -36.65 |
| Consensus MFE | -21.44 |
| Energy contribution | -24.28 |
| Covariance contribution | 2.83 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19828230 107 + 22224390 CGCCUGUCAGCUGGCAGU--UUUCCUUUACCAGCUGGCAGUGCC-UGGCUGACGUGAAUUCCCGAAAAGUCACAGGGCCUACAAUGGCAGUGAAAAAUUGAGUUAGCGAU .(((((((((((((.((.--......)).))))))))))).)).-..((((((((((.((......)).))))...(((......))).............))))))... ( -38.70) >DroPse_CAF1 29320 97 + 1 ---------GCUGGGUGUGAUGUCAGAUGUGAGAUGGGCG---A-UGGGUGAUGGGAAUUCCCAACAAGUCACAGGGCCUACAAUGGCAGUGAAAAAUUGAGUUAGCGAU ---------(((((.(.((((.(((..(((.(..(((((.---.-((..((.((((....)))).)).....))..)))))...).))).)))...))))).)))))... ( -26.80) >DroSim_CAF1 32137 104 + 1 CGCCUGUCAGCUGGCAGU--UUUCCUUUGCCAGCUGGCAG---C-CGGCUGACGGGAAUUCCCGAAAAGUCACAGGGCCUACAAUGGCAGUGAAAAAUUGAGUUAGCGAU ((.((((((((((((((.--......))))))))))))))---.-))(((((((((....)))......((((...(((......))).))))........))))))... ( -50.90) >DroEre_CAF1 24017 103 + 1 CGCCUGUCAUCUGGCAGU--UU-CCCCCACCAGCUGGCAG---C-CGGCUGACGGGAGUUCCCGAAAAGUCACAGGGCCUACAAUGGCAUUGAAAAAUUGAGUUAGCGAU ((.((((((.((((..(.--..-...)..)))).))))))---.-))(((((((((....))).........(((.(((......))).))).........))))))... ( -35.20) >DroYak_CAF1 24894 105 + 1 CGCCUGUCAGCUGGCUGU--UUUCCUUUGCCAGCUGGCAG---CCCGGCUGACGGGAAUUCCCGAAAAGUCACAGGGCCUACAAUGGCAGUGAAAAAUUGAGUUAGCGAU ((.((((((((((((...--........))))))))))))---..))(((((((((....)))......((((...(((......))).))))........))))))... ( -46.30) >DroPer_CAF1 29954 97 + 1 ---------GCUGGGUGUGAUGUCAGAUGUGAGAUGGGAG---A-UGGGUGAAGGGAAUUCCCAACAAGUCACAGGGCCUACAAUGGCAGUGAAAAAUUGAGUUAGCGAU ---------(((((.(.((((.(((..(((.(..((((..---.-...((((.(((....)))......))))....))))...).))).)))...))))).)))))... ( -22.00) >consensus CGCCUGUCAGCUGGCAGU__UUUCCUUUGCCAGCUGGCAG___C_CGGCUGACGGGAAUUCCCGAAAAGUCACAGGGCCUACAAUGGCAGUGAAAAAUUGAGUUAGCGAU ...((((((.((((...............)))).)))))).......(((((((((....)))......((((...(((......))).))))........))))))... (-21.44 = -24.28 + 2.83)

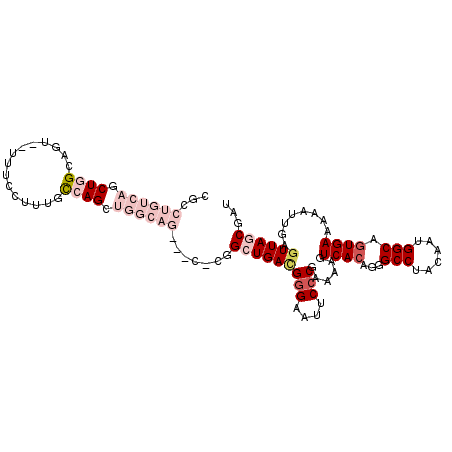
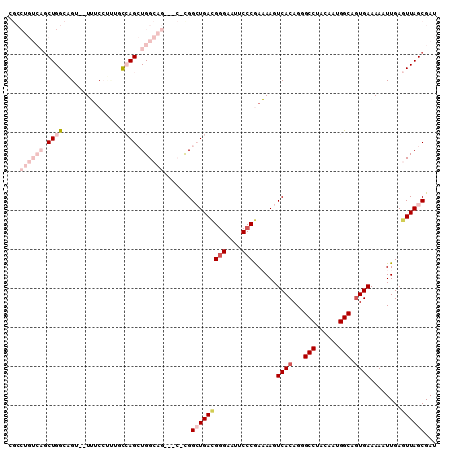
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:43 2006