
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,825,730 – 19,825,881 |
| Length | 151 |
| Max. P | 0.969429 |

| Location | 19,825,730 – 19,825,848 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.87 |
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -29.04 |
| Energy contribution | -29.24 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19825730 118 + 22224390 AGUUUGGCUUGAUGUUUGUCUUUUGUUUGUUGACACAAUGUUCGCGGAAAGGCG--AGUUGGCUAAAGCCGAAUGCAAAGAGGACUAAUUCAACAUUUUAGUCAAGAUGAAAAUUAACAU ..(((((((.((((((.(((((((.(((((.(((.....))).)))))...(((--..(((((....))))).)))..)))))))......))))))..))))))).............. ( -28.60) >DroSec_CAF1 31488 118 + 1 AGUUUGGCUUGGUGUUUGUCUUUUGUUUGUUGACACAAUGUUCGCGGGAAGGCG--AGUUGGCUAAAGCCGAAUGCAAAGAGGACUAAUUAAACAUUUUAGCCAAGAUGAAAAUGAACAU ..(((((((.(((((((((((((..(((((..........(((((......)))--))(((((....)))))..))))))))))).....)))))))..))))))).............. ( -31.40) >DroSim_CAF1 29654 118 + 1 AGUUUGGCUUGCUGUUUGUCUUUUGUUUGUUGACACAAUGUUCGCGGGAAGGCG--AGUUGGCUAAAGCCGAAUGCAAAGAGGACUAAUUAAACAUUUUAGCCAAGAUGAAAAUGAACAU ..(((((((...(((((((((((..(((((..........(((((......)))--))(((((....)))))..))))))))))).....)))))....))))))).............. ( -28.80) >DroEre_CAF1 21779 118 + 1 AGUUGGGCUUGGUGUUUGUCUUCUGUUUGUUGGCACAGUGUUCGCGGGAAGGCG--AGUUGGCUGAAGCCGAAUGCAAAUAGGACUAAUUAAACAUUUUAGCCAAGAUGAAAAUUAAGAU .....((((.((((((((..((((((((((((((.((((.(((((......)))--))...))))..))))...)))))))))).....))))))))..))))..(((....)))..... ( -35.50) >DroYak_CAF1 22624 120 + 1 AGUUUGGCUUGGUGUUUGUCUUUUGUUUGUUGACACAAUGUUCGCGGGAAGGCGAGAGUUGGCUAAAGCCGAAUGCAAAGAGGACUAAUUAAACAUUUUAGCCAAGAUGAAAAUGAACAU ..(((((((.(((((((((((((..(((((..........(((((......)))))..(((((....)))))..))))))))))).....)))))))..))))))).............. ( -31.60) >consensus AGUUUGGCUUGGUGUUUGUCUUUUGUUUGUUGACACAAUGUUCGCGGGAAGGCG__AGUUGGCUAAAGCCGAAUGCAAAGAGGACUAAUUAAACAUUUUAGCCAAGAUGAAAAUGAACAU ..(((((((.((((((((((((((.(((((.(((.....))).)))))...(((....(((((....))))).)))..))))))).....)))))))..))))))).............. (-29.04 = -29.24 + 0.20)


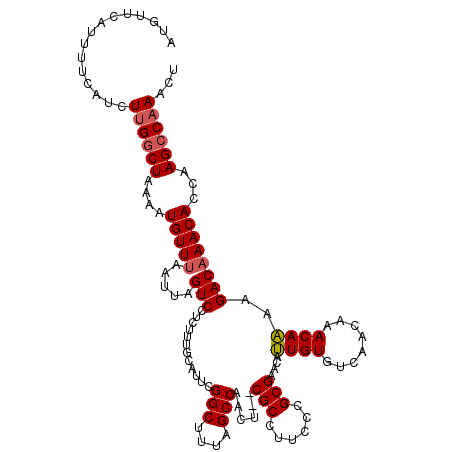
| Location | 19,825,730 – 19,825,848 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.87 |
| Mean single sequence MFE | -20.72 |
| Consensus MFE | -17.38 |
| Energy contribution | -17.66 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19825730 118 - 22224390 AUGUUAAUUUUCAUCUUGACUAAAAUGUUGAAUUAGUCCUCUUUGCAUUCGGCUUUAGCCAACU--CGCCUUUCCGCGAACAUUGUGUCAACAAACAAAAGACAAACAUCAAGCCAAACU .((((..........(((((...(((((((((((((......))).))))(((....)))....--(((......)))))))))..))))).........))))................ ( -18.51) >DroSec_CAF1 31488 118 - 1 AUGUUCAUUUUCAUCUUGGCUAAAAUGUUUAAUUAGUCCUCUUUGCAUUCGGCUUUAGCCAACU--CGCCUUCCCGCGAACAUUGUGUCAACAAACAAAAGACAAACACCAAGCCAAACU ...............((((((....(((((.....(((...((((.....(((....)))...(--(((......))))...((((....)))).)))).))))))))...))))))... ( -21.20) >DroSim_CAF1 29654 118 - 1 AUGUUCAUUUUCAUCUUGGCUAAAAUGUUUAAUUAGUCCUCUUUGCAUUCGGCUUUAGCCAACU--CGCCUUCCCGCGAACAUUGUGUCAACAAACAAAAGACAAACAGCAAGCCAAACU ...............((((((....(((((.....(((...((((.....(((....)))...(--(((......))))...((((....)))).)))).))))))))...))))))... ( -22.30) >DroEre_CAF1 21779 118 - 1 AUCUUAAUUUUCAUCUUGGCUAAAAUGUUUAAUUAGUCCUAUUUGCAUUCGGCUUCAGCCAACU--CGCCUUCCCGCGAACACUGUGCCAACAAACAGAAGACAAACACCAAGCCCAACU .................((((....(((((.....(((............(((....)))...(--(((......))))...((((........))))..))))))))...))))..... ( -20.40) >DroYak_CAF1 22624 120 - 1 AUGUUCAUUUUCAUCUUGGCUAAAAUGUUUAAUUAGUCCUCUUUGCAUUCGGCUUUAGCCAACUCUCGCCUUCCCGCGAACAUUGUGUCAACAAACAAAAGACAAACACCAAGCCAAACU ...............((((((....(((((.....(((...((((.....(((....))).....((((......))))...((((....)))).)))).))))))))...))))))... ( -21.20) >consensus AUGUUCAUUUUCAUCUUGGCUAAAAUGUUUAAUUAGUCCUCUUUGCAUUCGGCUUUAGCCAACU__CGCCUUCCCGCGAACAUUGUGUCAACAAACAAAAGACAAACACCAAGCCAAACU ...............((((((....(((((.....(((............(((....)))......(((......)))....((((........))))..))))))))...))))))... (-17.38 = -17.66 + 0.28)


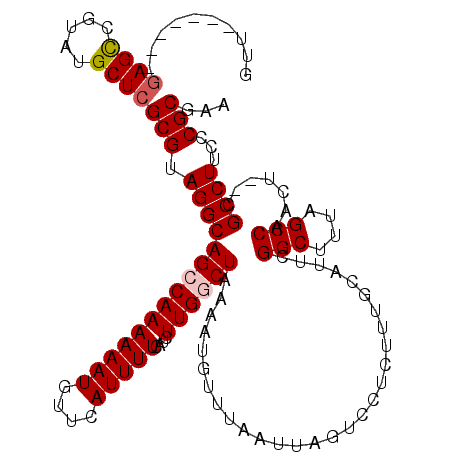
| Location | 19,825,770 – 19,825,881 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.19 |
| Mean single sequence MFE | -31.49 |
| Consensus MFE | -27.18 |
| Energy contribution | -27.26 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19825770 111 + 22224390 UUCGCGGAAAGGCG--AGUUGGCUAAAGCCGAAUGCAAAGAGGACUAAUUCAACAUUUUAGUCAAGAUGAAAAUUAACAUUUUUUGGCUGCCUACGCGAGCAUACGGCUC-------AAC ...(((...(((((--..(((((....)))))...(((((((((((((.........))))))..(((....)))....)))))))..))))).)))((((.....))))-------... ( -32.60) >DroSec_CAF1 31528 111 + 1 UUCGCGGGAAGGCG--AGUUGGCUAAAGCCGAAUGCAAAGAGGACUAAUUAAACAUUUUAGCCAAGAUGAAAAUGAACAUUUUUUGGCUGCCUACGCGAGCAUACGGCUC-------AAC ...(((...(((((--..(((((....)))))...).......................((((((((((........))))..)))))))))).)))((((.....))))-------... ( -32.20) >DroSim_CAF1 29694 111 + 1 UUCGCGGGAAGGCG--AGUUGGCUAAAGCCGAAUGCAAAGAGGACUAAUUAAACAUUUUAGCCAAGAUGAAAAUGAACAUUUUUUGGCUGCCUACGCGAGCAUACGGCUC-------AAC ...(((...(((((--..(((((....)))))...).......................((((((((((........))))..)))))))))).)))((((.....))))-------... ( -32.20) >DroEre_CAF1 21819 111 + 1 UUCGCGGGAAGGCG--AGUUGGCUGAAGCCGAAUGCAAAUAGGACUAAUUAAACAUUUUAGCCAAGAUGAAAAUUAAGAUUUUUUGGCUGCCUACGCGAGCAUACGACUC-------AAC ((((((...(((((--..(((((....)))))...).......................((((((((..............)))))))))))).))))))..........-------... ( -29.84) >DroYak_CAF1 22664 120 + 1 UUCGCGGGAAGGCGAGAGUUGGCUAAAGCCGAAUGCAAAGAGGACUAAUUAAACAUUUUAGCCAAGAUGAAAAUGAACAUUUUUUGCCUGCCUACGCGAGCAUACGGCUGAAAUACGAAC .(((((((.((((((((((((((....))))).........((.((((.........)))))).................))))))))).))..)))))((.....))............ ( -30.60) >consensus UUCGCGGGAAGGCG__AGUUGGCUAAAGCCGAAUGCAAAGAGGACUAAUUAAACAUUUUAGCCAAGAUGAAAAUGAACAUUUUUUGGCUGCCUACGCGAGCAUACGGCUC_______AAC ((((((...((((.....(((((....)))))...........................((((((((..............)))))))))))).)))))).................... (-27.18 = -27.26 + 0.08)



| Location | 19,825,770 – 19,825,881 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.19 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -23.46 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19825770 111 - 22224390 GUU-------GAGCCGUAUGCUCGCGUAGGCAGCCAAAAAAUGUUAAUUUUCAUCUUGACUAAAAUGUUGAAUUAGUCCUCUUUGCAUUCGGCUUUAGCCAACU--CGCCUUUCCGCGAA ...-------((((.....))))(((.((((.((...(((((....)))))......((((((.........))))))......))....(((....)))....--.))))...)))... ( -25.50) >DroSec_CAF1 31528 111 - 1 GUU-------GAGCCGUAUGCUCGCGUAGGCAGCCAAAAAAUGUUCAUUUUCAUCUUGGCUAAAAUGUUUAAUUAGUCCUCUUUGCAUUCGGCUUUAGCCAACU--CGCCUUCCCGCGAA (((-------((((((.((((..((....))((((((...(((........))).)))))).......................)))).)))).)))))....(--(((......)))). ( -28.50) >DroSim_CAF1 29694 111 - 1 GUU-------GAGCCGUAUGCUCGCGUAGGCAGCCAAAAAAUGUUCAUUUUCAUCUUGGCUAAAAUGUUUAAUUAGUCCUCUUUGCAUUCGGCUUUAGCCAACU--CGCCUUCCCGCGAA (((-------((((((.((((..((....))((((((...(((........))).)))))).......................)))).)))).)))))....(--(((......)))). ( -28.50) >DroEre_CAF1 21819 111 - 1 GUU-------GAGUCGUAUGCUCGCGUAGGCAGCCAAAAAAUCUUAAUUUUCAUCUUGGCUAAAAUGUUUAAUUAGUCCUAUUUGCAUUCGGCUUCAGCCAACU--CGCCUUCCCGCGAA (((-------((((((.((((....(((((((((((((((((....)))))....))))))..(((.....))).).)))))..)))).)))).)))))....(--(((......)))). ( -29.10) >DroYak_CAF1 22664 120 - 1 GUUCGUAUUUCAGCCGUAUGCUCGCGUAGGCAGGCAAAAAAUGUUCAUUUUCAUCUUGGCUAAAAUGUUUAAUUAGUCCUCUUUGCAUUCGGCUUUAGCCAACUCUCGCCUUCCCGCGAA .(((((......(((.((((....)))))))((((..(((((....)))))....(((((((((..((..((((((......))).)))..))))))))))).....))))....))))) ( -27.30) >consensus GUU_______GAGCCGUAUGCUCGCGUAGGCAGCCAAAAAAUGUUCAUUUUCAUCUUGGCUAAAAUGUUUAAUUAGUCCUCUUUGCAUUCGGCUUUAGCCAACU__CGCCUUCCCGCGAA ..........((((.....))))(((.(((((((((((((((....)))))....)))))).............................(((....))).......))))...)))... (-23.46 = -23.90 + 0.44)


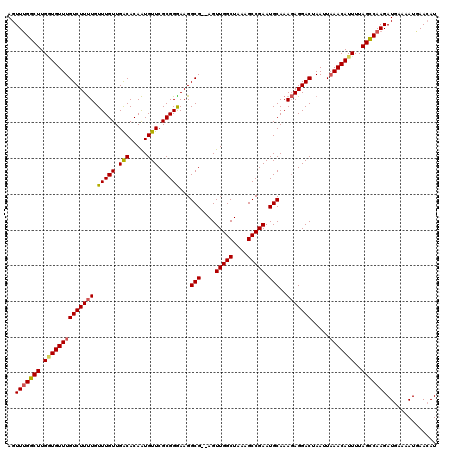
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:33 2006