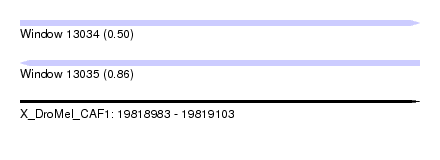
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,818,983 – 19,819,103 |
| Length | 120 |
| Max. P | 0.858891 |

| Location | 19,818,983 – 19,819,103 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.12 |
| Mean single sequence MFE | -28.04 |
| Consensus MFE | -14.31 |
| Energy contribution | -14.97 |
| Covariance contribution | 0.66 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.51 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19818983 120 + 22224390 CGGGUAAGUUGACCGGAAGUGAUUAUCUACCAAAAUCGGUGCUUAGACAUGGUCAACUAAUAAGCCUAGUGUAUGCUGGUGUAUUUUGUAUAAUUAACCAGCAUCUUUGCACCAUUUAAU .((((.((((((((....(((.(((..((((......))))..))).))))))))))).....)))).((((((((((((.(((.....)))....))))))))....))))........ ( -34.20) >DroSec_CAF1 24860 108 + 1 UGGUUAAGUUGACCGGAAGUGAUUAUCUACCAAAAUCGGUGCUUA----------AGUAAUAAGCCUAA--UAUGCCGGUGUAUUUUGGAUAAUUAACCAGCAUCUUUGCUCCAUUAAAU .((((.....))))(((..((((((((((.....(((((((((((----------.....)))))....--...))))))......))))))))))....(((....))))))....... ( -27.01) >DroSim_CAF1 21618 108 + 1 UGGUUAAGUUGACCGGAAGUGAUUAUCUACCAAAAUCGGUGCUUA----------AGUAAUAAGCCUAA--UAUGCCGGUGUAUUUUGGAUAAUUAACCAGCAUCUUUGCUCCAUUAAAU .((((.....))))(((..((((((((((.....(((((((((((----------.....)))))....--...))))))......))))))))))....(((....))))))....... ( -27.01) >DroEre_CAF1 15233 101 + 1 UAGGUAAAUUGACCAAAAG-----------------UGGUGCUUCGACUUGGUCACGCAAUAAGCCUAA--UAUGCUGGUGUAUUUUGGACAAAUAACCAGCAUCUUUGCACAAUUUAAU ....(((((((((((..((-----------------(.(.....).)))))))).........((..((--.((((((((.(((((.....))))))))))))).)).))..)))))).. ( -25.10) >DroYak_CAF1 15690 118 + 1 UGGGUGAGUUGACCGAAAGUGAUUAUUUAACAAAAUUGGUGCUUAGACGUGGUCAAGUAAUAAGCCUAU--UAUGCUGGUGUAUUUUGGACAAUUAACCAGCAUCUUUGCACCGUUUAAU ((..(((((...((....).)...))))).)).....(((((..(((...(((..(....)..)))...--..(((((((................))))))))))..)))))....... ( -26.89) >consensus UGGGUAAGUUGACCGGAAGUGAUUAUCUACCAAAAUCGGUGCUUAGAC_UGGUCAAGUAAUAAGCCUAA__UAUGCUGGUGUAUUUUGGAUAAUUAACCAGCAUCUUUGCACCAUUUAAU .(((((((...(((((...((.........))...)))))(((((...............))))).......((((((((................)))))))).))))).))....... (-14.31 = -14.97 + 0.66)


| Location | 19,818,983 – 19,819,103 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.12 |
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -15.04 |
| Energy contribution | -15.56 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19818983 120 - 22224390 AUUAAAUGGUGCAAAGAUGCUGGUUAAUUAUACAAAAUACACCAGCAUACACUAGGCUUAUUAGUUGACCAUGUCUAAGCACCGAUUUUGGUAGAUAAUCACUUCCGGUCAACUUACCCG ......(((((.....((((((((..(((......)))..)))))))).)))))((......((((((((..........((((....)))).((........)).))))))))...)). ( -29.30) >DroSec_CAF1 24860 108 - 1 AUUUAAUGGAGCAAAGAUGCUGGUUAAUUAUCCAAAAUACACCGGCAUA--UUAGGCUUAUUACU----------UAAGCACCGAUUUUGGUAGAUAAUCACUUCCGGUCAACUUAACCA .......((((((....)))((((((.(((.(((((((....(((....--....(((((.....----------))))).))))))))))))).))))))..)))(((.......))). ( -22.30) >DroSim_CAF1 21618 108 - 1 AUUUAAUGGAGCAAAGAUGCUGGUUAAUUAUCCAAAAUACACCGGCAUA--UUAGGCUUAUUACU----------UAAGCACCGAUUUUGGUAGAUAAUCACUUCCGGUCAACUUAACCA .......((((((....)))((((((.(((.(((((((....(((....--....(((((.....----------))))).))))))))))))).))))))..)))(((.......))). ( -22.30) >DroEre_CAF1 15233 101 - 1 AUUAAAUUGUGCAAAGAUGCUGGUUAUUUGUCCAAAAUACACCAGCAUA--UUAGGCUUAUUGCGUGACCAAGUCGAAGCACCA-----------------CUUUUGGUCAAUUUACCUA ..(((((((.((....(((((((((((((.....))))).)))))))).--(((.((.....)).)))..........))((((-----------------....))))))))))).... ( -25.40) >DroYak_CAF1 15690 118 - 1 AUUAAACGGUGCAAAGAUGCUGGUUAAUUGUCCAAAAUACACCAGCAUA--AUAGGCUUAUUACUUGACCACGUCUAAGCACCAAUUUUGUUAAAUAAUCACUUUCGGUCAACUCACCCA .......(((((....((((((((..(((......)))..)))))))).--.(((((...............))))).))))).......................(((......))).. ( -25.56) >consensus AUUAAAUGGUGCAAAGAUGCUGGUUAAUUAUCCAAAAUACACCAGCAUA__UUAGGCUUAUUACUUGACCA_GUCUAAGCACCGAUUUUGGUAGAUAAUCACUUCCGGUCAACUUACCCA .......((.((....((((((((..(((......)))..)))))))).......)).....................(.((((....((((.....))))....))))).......)). (-15.04 = -15.56 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:25 2006