
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,814,238 – 19,814,369 |
| Length | 131 |
| Max. P | 0.998781 |

| Location | 19,814,238 – 19,814,329 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 82.85 |
| Mean single sequence MFE | -22.40 |
| Consensus MFE | -18.95 |
| Energy contribution | -18.73 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19814238 91 - 22224390 AC--UUGCAAAGUCGGAGGAGGAGACCCGGGCA-AAUAGCUGGGAAUAAAUAAAGUCAAGGUAAGGACCUGCGACAAUCGCACACACACGAUCG ..--.......((((.(((.(....)((.(((.-....))).)).......................))).)))).((((........)))).. ( -21.50) >DroEre_CAF1 12483 84 - 1 AC--UUGCAAAAC------CGAAGACCCA-GCAAACCAGCUGGGAAUAAAUAAAGUCAAGGUGAGGACCUGCGACAAUCGCACAUU-UCGAUCG ..--.........------(((((.((((-((......))))))..........(((.((((....))))..))).........))-))).... ( -23.40) >DroYak_CAF1 12799 88 - 1 ACUCUUGCAAGAG------CGGAGACCCAGGCAAACCAGCUGGGAAUAAAUAAAGUCAAGGUGAGGACCUGCGACAAUCGCACACACACGAUCG .((((.((....)------))))).(((((.(......))))))..........(((.((((....))))..))).((((........)))).. ( -22.30) >consensus AC__UUGCAAAAC______CGGAGACCCAGGCAAACCAGCUGGGAAUAAAUAAAGUCAAGGUGAGGACCUGCGACAAUCGCACACACACGAUCG .....(((.................((((.((......))))))..........(((.((((....))))..)))....)))............ (-18.95 = -18.73 + -0.22)


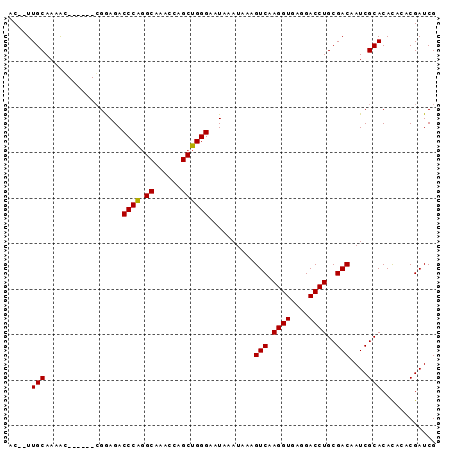
| Location | 19,814,254 – 19,814,369 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -28.14 |
| Consensus MFE | -17.34 |
| Energy contribution | -17.70 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19814254 115 + 22224390 AUUGUCGCAGGUCCUUACCUUGACUUUAUUUAUUCCCAGCUAUU-UGCCCGGGUCUCCUCCUCCGACUUUGCAA--GUGCUUGUUGGCAAGGCGAUUGCCUUUAUUUCUCUUAAGAGU ...((((.((((....))))))))...........(((((((((-(((..(((((.........))))).))))--)))...))))).(((((....))))).....(((....))). ( -27.20) >DroSec_CAF1 20234 92 + 1 AUUGUCGCAGGUCCUCACCUUGACUUUUUUUAUUCCCAGCUAUU-UGCCUGGGUCUCCUC---G----------------------GCAAGGCGAUUGCCUUUAUUUUUCUUAAGAGU ...((((.((((....))))))))..((((((..((((((....-.).))))).......---(----------------------((((.....)))))...........)))))). ( -21.70) >DroSim_CAF1 17242 99 + 1 AUUGUCGCAGGUCCUCACCUUGACUUUAUUUAUUCCCAGCUAUU-UGCCUGGGUCUCCUC---GGACUU---------------UGGCAAGGCGAUUGCCUUUAUUUUUCUUAAGAGU ...((((.((((....)))))))).......((((........(-((((..(((((....---))))).---------------.)))))(((....)))..............)))) ( -24.20) >DroEre_CAF1 12498 109 + 1 AUUGUCGCAGGUCCUCACCUUGACUUUAUUUAUUCCCAGCUGGUUUGC-UGGGUCUUCG------GUUUUGCAA--GUGUGUGUUGCCAAGGCGAUUGCCUGUAUUUGUCUUUAGAGU ......((((((..((.((((.............((((((......))-)))).....(------((..((((.--...))))..))))))).))..))))))............... ( -33.30) >DroYak_CAF1 12815 112 + 1 AUUGUCGCAGGUCCUCACCUUGACUUUAUUUAUUCCCAGCUGGUUUGCCUGGGUCUCCG------CUCUUGCAAGAGUGUGUGUUGCCAAGGCGAUUGGCUGUAUUUUUCUUAAGAGU ...((((.((((....))))))))............((((..(((.((((.((((.(((------((((....)))))).).)..))).)))))))..))))................ ( -34.30) >consensus AUUGUCGCAGGUCCUCACCUUGACUUUAUUUAUUCCCAGCUAUU_UGCCUGGGUCUCCUC_____ACUUUGCAA__GUG__UGUUGGCAAGGCGAUUGCCUUUAUUUUUCUUAAGAGU ...((((.((((....))))))))..........((((((......)).))))................................(((((.....))))).................. (-17.34 = -17.70 + 0.36)

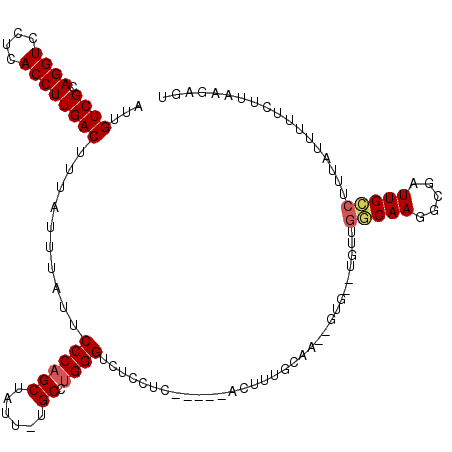
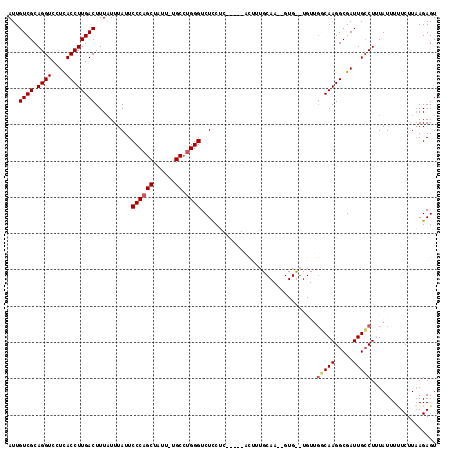
| Location | 19,814,254 – 19,814,369 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -30.36 |
| Consensus MFE | -19.56 |
| Energy contribution | -19.56 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.64 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19814254 115 - 22224390 ACUCUUAAGAGAAAUAAAGGCAAUCGCCUUGCCAACAAGCAC--UUGCAAAGUCGGAGGAGGAGACCCGGGCA-AAUAGCUGGGAAUAAAUAAAGUCAAGGUAAGGACCUGCGACAAU .(((((..((........(((((.....))))).....((..--..))....))..)))))....(((((...-.....)))))..........(((.((((....))))..)))... ( -30.30) >DroSec_CAF1 20234 92 - 1 ACUCUUAAGAAAAAUAAAGGCAAUCGCCUUGC----------------------C---GAGGAGACCCAGGCA-AAUAGCUGGGAAUAAAAAAAGUCAAGGUGAGGACCUGCGACAAU .(((((..(.......(((((....))))).)----------------------.---.))))).(((((...-.....)))))..........(((.((((....))))..)))... ( -25.70) >DroSim_CAF1 17242 99 - 1 ACUCUUAAGAAAAAUAAAGGCAAUCGCCUUGCCA---------------AAGUCC---GAGGAGACCCAGGCA-AAUAGCUGGGAAUAAAUAAAGUCAAGGUGAGGACCUGCGACAAU .(((((..((......(((((....)))))....---------------...)).---.))))).(((((...-.....)))))..........(((.((((....))))..)))... ( -29.42) >DroEre_CAF1 12498 109 - 1 ACUCUAAAGACAAAUACAGGCAAUCGCCUUGGCAACACACAC--UUGCAAAAC------CGAAGACCCA-GCAAACCAGCUGGGAAUAAAUAAAGUCAAGGUGAGGACCUGCGACAAU ................((((...((((((((((........(--((.(.....------.)))).((((-((......))))))..........))))))))))...))))....... ( -34.00) >DroYak_CAF1 12815 112 - 1 ACUCUUAAGAAAAAUACAGCCAAUCGCCUUGGCAACACACACUCUUGCAAGAG------CGGAGACCCAGGCAAACCAGCUGGGAAUAAAUAAAGUCAAGGUGAGGACCUGCGACAAU ................(((....((((((((((........((((.((....)------))))).(((((.(......))))))..........))))))))))....)))....... ( -32.40) >consensus ACUCUUAAGAAAAAUAAAGGCAAUCGCCUUGCCAACA__CAC__UUGCAAAAU_____GAGGAGACCCAGGCA_AAUAGCUGGGAAUAAAUAAAGUCAAGGUGAGGACCUGCGACAAU ..................(((((.....)))))................................((((.((......))))))..........(((.((((....))))..)))... (-19.56 = -19.56 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:22 2006