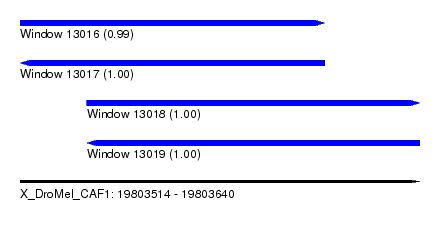
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,803,514 – 19,803,640 |
| Length | 126 |
| Max. P | 0.999292 |

| Location | 19,803,514 – 19,803,610 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 72.86 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -16.96 |
| Energy contribution | -17.88 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19803514 96 + 22224390 ----GAAAAAACAGAUAUGCAGACCGAAAAGUGC--AUAGACGAAAAGCAAAGCACAAGCGUUUGCAUUUUCUGCUUUUGCGCGUCUGCUUUUGCUUUUGGC--- ----..((((.((((...((((((......((((--(.((..((((((((((((....)).))))).)))))..))..))))))))))).)))).))))...--- ( -28.50) >DroSec_CAF1 2225 88 + 1 CAGCGGAAAAGCAGAUAUGAAA--------------AUCAGCCAAAAGCAAAGCACAAGCGUU-GGAUUUUCUGCUUUUGCGCGUCUACUUUUGCUUUUUGCU-- .((((.(((((((((...((..--------------....(((((((((...((....))...-((.....))))))))).)).))....)))))))))))))-- ( -24.10) >DroSim_CAF1 2294 102 + 1 CAGCGGAAAAGCAGAUAUGCAGACCCAAAAGUGCACAUAGGCCAAAAGCAAAGCACAAGCGUU-GGAUUUUCUGCUUUUGCGCGUCUACUUUUGCUUUUUGCU-- .((((((((.(((((...(((((.((((..((((......((.....))...)))).....))-))....))))).)))))(((........)))))))))))-- ( -27.90) >DroEre_CAF1 2028 79 + 1 ----------------------AUGGAAAAGUAU--AAAAGCCAAAAGCAAAGCACAAGCGUU-GGAUUUUCUGCUUUUGCGCGUCUGUUUUUGCUUUU-GCUUU ----------------------............--.(((((.((((((((((.(((..(((.-((.....))((....)))))..)))))))))))))-))))) ( -21.80) >DroYak_CAF1 2269 90 + 1 ------------AUGCAUGCAGAUGGAAAAGUGC--AUAAGCCAACAGCAAAGCACAAGCGUU-GGAUUUUCUGCUUUUGCGCGUCUGCUUUUGCUUUUCGCUUU ------------..(((.((((((((((((((..--.....(((((.((.........)))))-)).......)))))).).)))))))...))).......... ( -26.04) >consensus ____G_AAAA_CAGAUAUGCAGACCGAAAAGUGC__AUAAGCCAAAAGCAAAGCACAAGCGUU_GGAUUUUCUGCUUUUGCGCGUCUGCUUUUGCUUUUUGCU__ .......................................(((.((((((((((((...((((..((........))...))))...))).))))))))).))).. (-16.96 = -17.88 + 0.92)


| Location | 19,803,514 – 19,803,610 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 72.86 |
| Mean single sequence MFE | -24.74 |
| Consensus MFE | -16.78 |
| Energy contribution | -17.38 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19803514 96 - 22224390 ---GCCAAAAGCAAAAGCAGACGCGCAAAAGCAGAAAAUGCAAACGCUUGUGCUUUGCUUUUCGUCUAU--GCACUUUUCGGUCUGCAUAUCUGUUUUUUC---- ---...(((((((...((((((.((.(((((..(((((.(((((.((....))))))))))))((....--)).))))))))))))).....)))))))..---- ( -27.20) >DroSec_CAF1 2225 88 - 1 --AGCAAAAAGCAAAAGUAGACGCGCAAAAGCAGAAAAUCC-AACGCUUGUGCUUUGCUUUUGGCUGAU--------------UUUCAUAUCUGCUUUUCCGCUG --(((......((((((((((.(((((..(((.........-...))))))))))))))))))((.(((--------------......))).))......))). ( -24.20) >DroSim_CAF1 2294 102 - 1 --AGCAAAAAGCAAAAGUAGACGCGCAAAAGCAGAAAAUCC-AACGCUUGUGCUUUGCUUUUGGCCUAUGUGCACUUUUGGGUCUGCAUAUCUGCUUUUCCGCUG --(((.(((((((((((((((.(((((..(((.........-...)))))))))))))))))((((((..........)))))).........))))))..))). ( -29.60) >DroEre_CAF1 2028 79 - 1 AAAGC-AAAAGCAAAAACAGACGCGCAAAAGCAGAAAAUCC-AACGCUUGUGCUUUGCUUUUGGCUUUU--AUACUUUUCCAU---------------------- (((((-(((((((((.......(((((..(((.........-...))))))))))))))))).))))).--............---------------------- ( -18.11) >DroYak_CAF1 2269 90 - 1 AAAGCGAAAAGCAAAAGCAGACGCGCAAAAGCAGAAAAUCC-AACGCUUGUGCUUUGCUGUUGGCUUAU--GCACUUUUCCAUCUGCAUGCAU------------ ...((...((((...((((((.(((((..(((.........-...))))))))))))))....))))((--(((..........)))))))..------------ ( -24.60) >consensus __AGCAAAAAGCAAAAGCAGACGCGCAAAAGCAGAAAAUCC_AACGCUUGUGCUUUGCUUUUGGCUUAU__GCACUUUUCCAUCUGCAUAUCUG_UUUU_C____ ........(((((((((((((.(((((..(((.............))))))))))))))))).))))...................................... (-16.78 = -17.38 + 0.60)


| Location | 19,803,535 – 19,803,640 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.98 |
| Mean single sequence MFE | -38.84 |
| Consensus MFE | -25.28 |
| Energy contribution | -26.20 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -4.26 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19803535 105 + 22224390 GAAAAGUGC--AUAGACGAAAAGCAAAGCACAAGCGUUUGCAUUUUCUGCUUUUGCGCGUCUGCUUUUGCUUUUGGC-------------UUUUGCGCUGUUGCGCUUUUGCUUUCGCAC .....((((--.......((((((((((((...((((..(((.....)))....))))...))).)))))))))(((-------------....((((....))))....)))...)))) ( -36.40) >DroSec_CAF1 2247 101 + 1 -----------AUCAGCCAAAAGCAAAGCACAAGCGUU-GGAUUUUCUGCUUUUGCGCGUCUACUUUUGCUUUUUGCU-------UUUGCUUUUGCGCUGUUGCGCUUUUGCUUCCGCAC -----------..(((((((((((((((((.(((((.(-((((....(((....))).)))))....)))))..))).-------)))))))))).)))).((((..........)))). ( -31.60) >DroSim_CAF1 2319 112 + 1 CAAAAGUGCACAUAGGCCAAAAGCAAAGCACAAGCGUU-GGAUUUUCUGCUUUUGCGCGUCUACUUUUGCUUUUUGCU-------UUUGCUUUUGCGCUGUUGCGCUUUUGCUUCCGCAU ((((((((((....((((((((((((((((.(((((.(-((((....(((....))).)))))....)))))..))).-------)))))))))).)))..))))))))))......... ( -39.90) >DroEre_CAF1 2031 116 + 1 GAAAAGUAU--AAAAGCCAAAAGCAAAGCACAAGCGUU-GGAUUUUCUGCUUUUGCGCGUCUGUUUUUGCUUUU-GCUUUUGGCUUUGGCUUUUGCGCUGUUGCGCUUUUGCUUCCGCAC .((((((..--.((((((((((((((((((.(((((..-........))))).)))(((........))).)))-)))))))))))).))))))((((....))))...(((....))). ( -44.10) >DroYak_CAF1 2282 117 + 1 GAAAAGUGC--AUAAGCCAACAGCAAAGCACAAGCGUU-GGAUUUUCUGCUUUUGCGCGUCUGCUUUUGCUUUUCGCUUUUGCUUUUGGCUUUUGCGCUGUUGCGCUUUUGCUUCCGCAC ....(((((--(.(((((((.(((((((((.(((((..-........))))).)))(((...((....))....))).)))))).))))))).))))))..((((..........)))). ( -42.20) >consensus GAAAAGUGC__AUAAGCCAAAAGCAAAGCACAAGCGUU_GGAUUUUCUGCUUUUGCGCGUCUGCUUUUGCUUUUUGCU_______UUGGCUUUUGCGCUGUUGCGCUUUUGCUUCCGCAC ..............(((.((((((((((((...((((..((........))...))))...))).))))))))).)))................((((....))))...(((....))). (-25.28 = -26.20 + 0.92)



| Location | 19,803,535 – 19,803,640 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.98 |
| Mean single sequence MFE | -37.08 |
| Consensus MFE | -29.40 |
| Energy contribution | -30.00 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.07 |
| Mean z-score | -4.70 |
| Structure conservation index | 0.79 |
| SVM decision value | 3.49 |
| SVM RNA-class probability | 0.999292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19803535 105 - 22224390 GUGCGAAAGCAAAAGCGCAACAGCGCAAAA-------------GCCAAAAGCAAAAGCAGACGCGCAAAAGCAGAAAAUGCAAACGCUUGUGCUUUGCUUUUCGUCUAU--GCACUUUUC (((((...((....((((....))))....-------------((.....))....))((((((......)).(((((.(((((.((....)))))))))))))))).)--))))..... ( -34.10) >DroSec_CAF1 2247 101 - 1 GUGCGGAAGCAAAAGCGCAACAGCGCAAAAGCAAA-------AGCAAAAAGCAAAAGUAGACGCGCAAAAGCAGAAAAUCC-AACGCUUGUGCUUUGCUUUUGGCUGAU----------- .(((....))).........((((.((((((((((-------.(((..((((....(....)((......)).........-...)))).)))))))))))))))))..----------- ( -33.30) >DroSim_CAF1 2319 112 - 1 AUGCGGAAGCAAAAGCGCAACAGCGCAAAAGCAAA-------AGCAAAAAGCAAAAGUAGACGCGCAAAAGCAGAAAAUCC-AACGCUUGUGCUUUGCUUUUGGCCUAUGUGCACUUUUG .(((....)))...(((((...((.((((((((((-------.(((..((((....(....)((......)).........-...)))).)))))))))))))))...)))))....... ( -36.20) >DroEre_CAF1 2031 116 - 1 GUGCGGAAGCAAAAGCGCAACAGCGCAAAAGCCAAAGCCAAAAGC-AAAAGCAAAAACAGACGCGCAAAAGCAGAAAAUCC-AACGCUUGUGCUUUGCUUUUGGCUUUU--AUACUUUUC .(((....)))...((((....))))(((((..((((((((((((-(((.((..........))(((.((((.........-...)))).)))))))))))))))))).--...))))). ( -41.00) >DroYak_CAF1 2282 117 - 1 GUGCGGAAGCAAAAGCGCAACAGCGCAAAAGCCAAAAGCAAAAGCGAAAAGCAAAAGCAGACGCGCAAAAGCAGAAAAUCC-AACGCUUGUGCUUUGCUGUUGGCUUAU--GCACUUUUC (((((.........((((....))))..(((((((.((((((.(((....((....))...)))(((.((((.........-...)))).))))))))).))))))).)--))))..... ( -40.80) >consensus GUGCGGAAGCAAAAGCGCAACAGCGCAAAAGCAAA_______AGCAAAAAGCAAAAGCAGACGCGCAAAAGCAGAAAAUCC_AACGCUUGUGCUUUGCUUUUGGCUUAU__GCACUUUUC .(((....)))...((((....))))......................(((((((((((((.(((((..(((.............))))))))))))))))).))))............. (-29.40 = -30.00 + 0.60)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:10 2006