
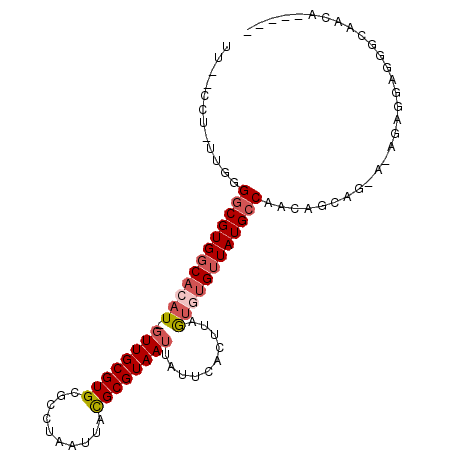
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,730,208 – 19,730,301 |
| Length | 93 |
| Max. P | 0.828139 |

| Location | 19,730,208 – 19,730,301 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -26.84 |
| Consensus MFE | -18.66 |
| Energy contribution | -19.47 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.828139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19730208 93 + 22224390 UU--CCU-UUGGGGCGUGGCACAU-GUUGCGUGCGCCUAAUUACGCGUAAUUAUUCACUUAGUGUGUUAUGACAACAGCAG-GAAAAGGAGGGGAACA----- ((--(((-(((..(((((((((((-((((((((..........))))))))..........)))))))))).)..))).))-))).............----- ( -25.80) >DroPse_CAF1 30102 87 + 1 UUUGUCU-UGGGGGCGUGGCACAU-GUUGCGUGCACUUAAUGAUGCGUAAUUAUUCACUUAGUUGGUUAUGCCACAAAAA-------UGAUGACGA------- .(((((.-...(.(((..((....-))..))).)..........(((((((((..........)))))))))........-------....)))))------- ( -20.50) >DroSec_CAF1 26047 93 + 1 UU--CCU-UUGGGGCGUGGCACAU-GUUGCGUGCGCCUAAUUCCGCGUAAUUAUUCACUUAGUGUGUUAUGCCAACAGCAG-AAAGAGGAGGGCAACA----- ((--(((-((..((((((((((((-((((((((..........))))))))..........))))))))))))........-..)))))))(....).----- ( -29.10) >DroSim_CAF1 2952 93 + 1 UU--CCU-UUGGGGCGUGGCACAU-GUUGCGUGCGCCUAAUUACGCGUAAUUAUUCACUUAGUGUGUUAUGCCAACAGCAG-AAAGAGGAGGGCAACA----- ((--(((-((..((((((((((((-((((((((..........))))))))..........))))))))))))........-..)))))))(....).----- ( -29.50) >DroYak_CAF1 23012 98 + 1 UU--CCUUUUGGGGCGUGGCACAU-GUUGCGUGCGCCUAAUUACGCGUAAUUAUUCACUUAGUGUGUUAUGCCAACAGCAG-C-AGAGAAGGGGAACAAAAAA ((--((((((..((((((((((((-((((((((..........))))))))..........))))))))))))........-.-...))))))))........ ( -29.04) >DroMoj_CAF1 32724 80 + 1 --------AGGGGGCGUGUCACGUUUUUGCGUGCACUUAAUUGUGCGUAAUUAUUCACUUUAGUUGUUAUGCCAACGAGUGGC-GGCCC-------------- --------...((((.((((((((..(((((..((......))..)))))..))........((((......))))..)))))-)))))-------------- ( -27.10) >consensus UU__CCU_UUGGGGCGUGGCACAU_GUUGCGUGCGCCUAAUUACGCGUAAUUAUUCACUUAGUGUGUUAUGCCAACAGCAG_A_AGAGGAGGGCAACA_____ ............((((((((((((.((((((((..........))))))))..........)))))))))))).............................. (-18.66 = -19.47 + 0.81)

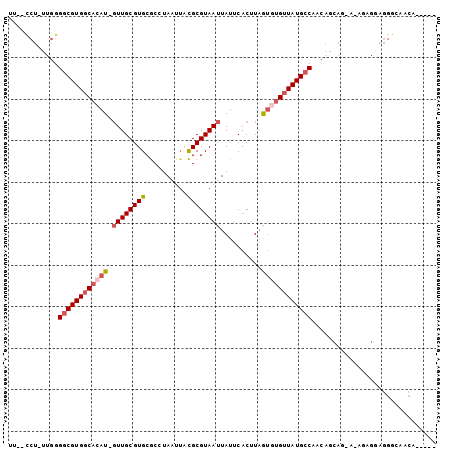
| Location | 19,730,208 – 19,730,301 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -21.06 |
| Consensus MFE | -12.64 |
| Energy contribution | -12.75 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19730208 93 - 22224390 -----UGUUCCCCUCCUUUUC-CUGCUGUUGUCAUAACACACUAAGUGAAUAAUUACGCGUAAUUAGGCGCACGCAAC-AUGUGCCACGCCCCAA-AGG--AA -----........((((((..-..((...........(((.....)))..((((((....))))))((((((......-.))))))..))...))-)))--). ( -19.10) >DroPse_CAF1 30102 87 - 1 -------UCGUCAUCA-------UUUUUGUGGCAUAACCAACUAAGUGAAUAAUUACGCAUCAUUAAGUGCACGCAAC-AUGUGCCACGCCCCCA-AGACAAA -------..(((....-------....(((((((((.........((((....))))((((......)))).......-.)))))))))......-.)))... ( -20.16) >DroSec_CAF1 26047 93 - 1 -----UGUUGCCCUCCUCUUU-CUGCUGUUGGCAUAACACACUAAGUGAAUAAUUACGCGGAAUUAGGCGCACGCAAC-AUGUGCCACGCCCCAA-AGG--AA -----........((((..((-(((((((((...)))))......((((....))))))))))...((((((......-.)))))).........-)))--). ( -20.90) >DroSim_CAF1 2952 93 - 1 -----UGUUGCCCUCCUCUUU-CUGCUGUUGGCAUAACACACUAAGUGAAUAAUUACGCGUAAUUAGGCGCACGCAAC-AUGUGCCACGCCCCAA-AGG--AA -----........((((....-..((.((.((((((.(((.....))).........((((......)))).......-.)))))))))).....-)))--). ( -21.40) >DroYak_CAF1 23012 98 - 1 UUUUUUGUUCCCCUUCUCU-G-CUGCUGUUGGCAUAACACACUAAGUGAAUAAUUACGCGUAAUUAGGCGCACGCAAC-AUGUGCCACGCCCCAAAAGG--AA ...........((((...(-(-..((.((.((((((.(((.....))).........((((......)))).......-.))))))))))..)).))))--.. ( -21.50) >DroMoj_CAF1 32724 80 - 1 --------------GGGCC-GCCACUCGUUGGCAUAACAACUAAAGUGAAUAAUUACGCACAAUUAAGUGCACGCAAAAACGUGACACGCCCCCU-------- --------------((((.-((((.....))))............(((.........((((......)))).(((......))).)))))))...-------- ( -23.30) >consensus _____UGUUGCCCUCCUCU_G_CUGCUGUUGGCAUAACACACUAAGUGAAUAAUUACGCGUAAUUAGGCGCACGCAAC_AUGUGCCACGCCCCAA_AGG__AA .............................(((((((.........((((....))))((((......)))).........)))))))................ (-12.64 = -12.75 + 0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:57 2006