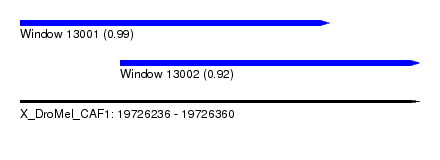
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,726,236 – 19,726,360 |
| Length | 124 |
| Max. P | 0.987006 |

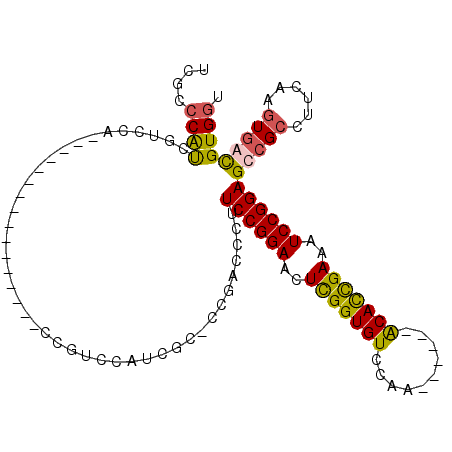
| Location | 19,726,236 – 19,726,332 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 70.64 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -21.52 |
| Energy contribution | -21.28 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.987006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19726236 96 + 22224390 UCGCCCAUCGUUCACCGUC---------CGCCGUCCAUCGCCCCGAUUCUUCCGGAACUCGGUGUCCAA------ACAUUGAAAUCCGGAGACGCCUUCAAGUGAUGUGGU ..((((((((..(((((..---------..(((...((((...)))).....)))....)))))..)..------((.(((((...((....))..))))))))))).))) ( -28.40) >DroSec_CAF1 22167 89 + 1 UCGCCCAUUGUCCA----------------CCGUUCGUCGCCCCGAUUCUUCCGGAACUCGGUGUCCAA------ACAUCGAAAUCCGGAGCCGCCUUCAAGUUAUGUGUU ............((----------------(...(((......)))...(((((((..(((((((....------)))))))..)))))))...............))).. ( -20.60) >DroEre_CAF1 18965 79 + 1 UCGCCCGUCGCCC-----------------------AU--C-CCUUCCCUUCCGGAACUUGGUGUCCAA------GCACCGAAAUCCGGAGCCGCCUUCAAGUGGCGUGGU ...........((-----------------------((--.-........((((((..(((((((....------)))))))..))))))(((((......))))))))). ( -30.20) >DroYak_CAF1 19049 110 + 1 UCGUCCACCAUCCACCAUCCACCACAUCCACCGUCCAUCGC-CCGACCCUUCCGGAACUUGGUGUCCAAACCGAAACACCGAAAUCCGGAGCCGCCUUCAAAUGGCGUGGU ....................(((((.......(((......-..)))..(((((((..(((((((..........)))))))..)))))))..(((.......)))))))) ( -30.50) >consensus UCGCCCAUCGUCCA________________CCGUCCAUCGC_CCGACCCUUCCGGAACUCGGUGUCCAA______ACACCGAAAUCCGGAGCCGCCUUCAAGUGACGUGGU ....((((..........................................((((((..(((((((..........)))))))..))))))(((((......))))))))). (-21.52 = -21.28 + -0.25)


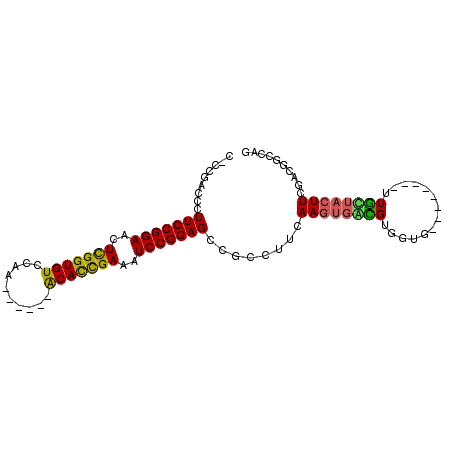
| Location | 19,726,267 – 19,726,360 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 76.95 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -20.05 |
| Energy contribution | -19.49 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19726267 93 + 22224390 CCCCGAUUCUUCCGGAACUCGGUGUCCAA------ACAUUGAAAUCCGGAGACGCCUUCAAGUGAUGUGGUGAUGUUGGGUUGUUACUUCGAUGGCCAG ...((((..(((((((..(((((((....------)))))))..))))))).((((.((....))...))))..))))(((..((.....))..))).. ( -29.10) >DroSec_CAF1 22191 85 + 1 CCCCGAUUCUUCCGGAACUCGGUGUCCAA------ACAUCGAAAUCCGGAGCCGCCUUCAAGUUAUGUGUUG--------UUACUUCUUCGACUGCCAG .........(((((((..(((((((....------)))))))..)))))))..((..(((((....(((...--------.)))..))).))..))... ( -22.00) >DroEre_CAF1 18980 84 + 1 C-CCUUCCCUUCCGGAACUUGGUGUCCAA------GCACCGAAAUCCGGAGCCGCCUUCAAGUGGCGUGGUG--------UUGCUACUUCCACUAGCAG .-........((((((..(((((((....------)))))))..))))))(((((......)))))((((.(--------(....))..))))...... ( -32.50) >DroYak_CAF1 19089 89 + 1 C-CCGACCCUUCCGGAACUUGGUGUCCAAACCGAAACACCGAAAUCCGGAGCCGCCUUCAAAUGGCGUGGUG--------UUGUUACUUCGACG-UCGG .-((((((((((((((..(((((((..........)))))))..))))))).((((.......)))).)).(--------(((......)))))-)))) ( -34.80) >consensus C_CCGACCCUUCCGGAACUCGGUGUCCAA______ACACCGAAAUCCGGAGCCGCCUUCAAGUGACGUGGUG________UUGCUACUUCGACGGCCAG .........(((((((..(((((((..........)))))))..)))))))........((((((((..............)))))))).......... (-20.05 = -19.49 + -0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:56 2006