
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,714,937 – 19,715,061 |
| Length | 124 |
| Max. P | 0.987120 |

| Location | 19,714,937 – 19,715,028 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 71.06 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -10.94 |
| Energy contribution | -12.03 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19714937 91 + 22224390 CAAAAGCGUUUAGCGGCAGUCAGAUUAUACUUGCGUUUAUGCUAAUGC----------U----------UUUAUGCUGCUCUGAUGACGAAUCAAUGUCAUCAAGGGUGGU .(((((((((.((((...((.((......)).)).....)))))))))----------)----------)))..((..(((((((((((......))))))).))))..)) ( -30.20) >DroSec_CAF1 11151 88 + 1 CAAAAGCGUUUAGCGGCAGUCAGAUUAUACUUGCGUUUAUGCUAAUGC----------U----------UUUAUGCUGCUCUGAUGACGAAUCAAUGUCAUCAAGGG---U .(((((((((.((((...((.((......)).)).....)))))))))----------)----------))).....((((((((((((......))))))).))))---) ( -25.80) >DroEre_CAF1 8075 88 + 1 CCAAAGCGUUUAGCGGCAGUCAGAUUAUACUUGCGUUUAUGCUAAUGC----------U----------UUUAUGCUGCCCUGAUGACGAAUCAAUGUCAUCAAGGU---A ..((((((((.((((...((.((......)).)).....)))))))))----------)----------)).....((((.((((((((......)))))))).)))---) ( -25.50) >DroWil_CAF1 11913 78 + 1 UA-AAGCGUUUUGCUGGAAAAGGAUUAUGUUUACACUUACAUUUGUGC----------U----------UAUUUAACGCAUUGACAGCCAAU----GGCAACG-------- ..-....(((.(((((((.(((.((.((((........))))..)).)----------)----------).))))..)))..))).(((...----)))....-------- ( -12.30) >DroYak_CAF1 7959 88 + 1 CAAAAGCGUUUAGCGGCAGUCAGAUUAUACUUGCGUUUAUGCUAAUGC----------U----------UUUAUGCUGCUCUGAUGACAAAUCAAUGUCAUCAAGGG---U .(((((((((.((((...((.((......)).)).....)))))))))----------)----------))).....((((((((((((......))))))).))))---) ( -27.00) >DroAna_CAF1 13196 104 + 1 CAAAAGCGUUUAGCGGCAGCCAGAUUAUGUUUGCGUUUAUGCUGAUGGCUGCUGCUGCUUCUCUUCCUCUUCUUGCUGCACU---GACGAGCCAAGGUCGUUAAGGG---- ...((((....(((((((((((...((((........))))....)))))))))))))))(((((.....(((((..((...---.....))))))).....)))))---- ( -29.80) >consensus CAAAAGCGUUUAGCGGCAGUCAGAUUAUACUUGCGUUUAUGCUAAUGC__________U__________UUUAUGCUGCUCUGAUGACGAAUCAAUGUCAUCAAGGG___U ............((((((((........))..(((((......))))).........................))))))..(((((((........)))))))........ (-10.94 = -12.03 + 1.09)
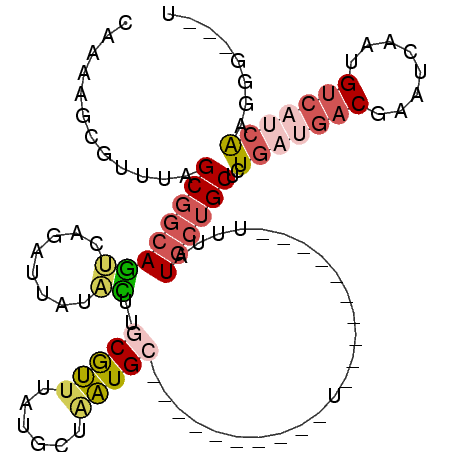
| Location | 19,714,968 – 19,715,061 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 81.64 |
| Mean single sequence MFE | -28.73 |
| Consensus MFE | -19.99 |
| Energy contribution | -19.43 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19714968 93 + 22224390 UGCGUUUAUGCUAAUGCUUUUAUGCUGCUCUGAUGACGAAUCAAUGUCAUCAAGGGUGGUGUGCAAUUGAGAGUGU-GGGAGUGGGGCGGACAA ..(((((...((.((((((((((((..(((((((((((......))))))).))))..))))......))))))))-.))....)))))..... ( -28.10) >DroSec_CAF1 11182 91 + 1 UGCGUUUAUGCUAAUGCUUUUAUGCUGCUCUGAUGACGAAUCAAUGUCAUCAAGGG---UGCGCAAAUGAGAGUGGCAGUGGUGGGGUGGGCAA ...((((((.(((.((((....((((((((((((((((......))))))).))))---)).))).((....))))))....))).)))))).. ( -28.50) >DroEre_CAF1 8106 76 + 1 UGCGUUUAUGCUAAUGCUUUUAUGCUGCCCUGAUGACGAAUCAAUGUCAUCAAGGU---AGUG---------------GGAGUGGGGUAGGCAA ...((((((.((...((((((..((((((.((((((((......)))))))).)))---))))---------------))))))).)))))).. ( -28.70) >DroYak_CAF1 7990 88 + 1 UGCGUUUAUGCUAAUGCUUUUAUGCUGCUCUGAUGACAAAUCAAUGUCAUCAAGGG---UGUGCAAGUGGCAUU---CGUAGUGGGGUGGACAA ...((((((.((((((((....((((((((((((((((......))))))).))))---)).)))...))))).---.....))).)))))).. ( -29.60) >consensus UGCGUUUAUGCUAAUGCUUUUAUGCUGCUCUGAUGACGAAUCAAUGUCAUCAAGGG___UGUGCAA_UGAGAGU____GGAGUGGGGUGGACAA ...((((((.(((..((......))..((.((((((((......)))))))).))...........................))).)))))).. (-19.99 = -19.43 + -0.56)

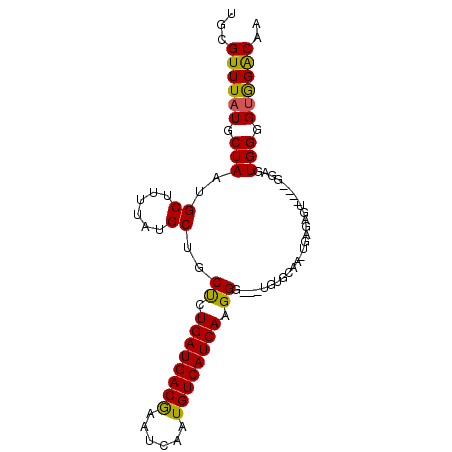

| Location | 19,714,968 – 19,715,061 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 81.64 |
| Mean single sequence MFE | -16.93 |
| Consensus MFE | -12.12 |
| Energy contribution | -11.88 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19714968 93 - 22224390 UUGUCCGCCCCACUCCC-ACACUCUCAAUUGCACACCACCCUUGAUGACAUUGAUUCGUCAUCAGAGCAGCAUAAAAGCAUUAGCAUAAACGCA .................-...........(((.........((((((((........))))))))....((......))....)))........ ( -13.70) >DroSec_CAF1 11182 91 - 1 UUGCCCACCCCACCACUGCCACUCUCAUUUGCGCA---CCCUUGAUGACAUUGAUUCGUCAUCAGAGCAGCAUAAAAGCAUUAGCAUAAACGCA .(((.(..........(((((........)).)))---....(((((((........)))))))).)))((......))....((......)). ( -16.30) >DroEre_CAF1 8106 76 - 1 UUGCCUACCCCACUCC---------------CACU---ACCUUGAUGACAUUGAUUCGUCAUCAGGGCAGCAUAAAAGCAUUAGCAUAAACGCA .(((............---------------..((---.((((((((((........)))))))))).)).......((....))......))) ( -19.00) >DroYak_CAF1 7990 88 - 1 UUGUCCACCCCACUACG---AAUGCCACUUGCACA---CCCUUGAUGACAUUGAUUUGUCAUCAGAGCAGCAUAAAAGCAUUAGCAUAAACGCA .................---(((((....(((...---.(.(((((((((......))))))))).)..))).....))))).((......)). ( -18.70) >consensus UUGCCCACCCCACUAC____ACUCUCA_UUGCACA___CCCUUGAUGACAUUGAUUCGUCAUCAGAGCAGCAUAAAAGCAUUAGCAUAAACGCA .(((.....................................((((((((........))))))))....((......))....)))........ (-12.12 = -11.88 + -0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:52 2006