| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,706,035 – 19,706,129 |
| Length | 94 |
| Max. P | 0.899942 |

| Location | 19,706,035 – 19,706,129 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -43.37 |
| Consensus MFE | -31.52 |
| Energy contribution | -31.11 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
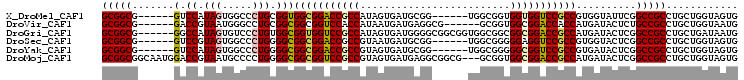
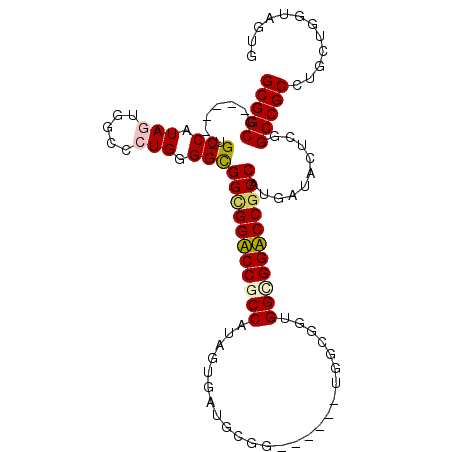
>X_DroMel_CAF1 19706035 94 + 22224390 CACUACCAGCAGGCGGCCGAAUACCACGGCGGACCACCACCGCCA------CCGCAUCACUAUGGCGGUCCGCCACCGCAGGGCCACUAUGGAC------CGCCGC ........((.(((((...........(((((.......))))).------........((((((.(((((((....)).))))).)))))).)------)))))) ( -37.70) >DroVir_CAF1 297 94 + 1 CAUUACCAGCAGGCGGCAGAGUAUCAUGGUGGUCCGCCACCGC------CGCCUCAUCAUUAUGGUGGACCGCCGCCGCAGGCCCAUUACGGUC------CGCCGC ........(((((((((.(.....)..(((((....)))))))------))))).........(((((((((..(((...)))......)))))------)))))) ( -43.00) >DroGri_CAF1 306 100 + 1 CAUUAUCAGCAGGCGGCCGAGUAUCAUGGCGGUCCGCCGCCGCCACCGCCGCCCCAUCACUAUGGCGGACCACCGCCACAGGGACACUAUGGCC------CGCCGC ........((.(((((((((((.((.(((((((.....)))))))..............((.((((((....)))))).)).)).))).))).)------)))))) ( -42.50) >DroSec_CAF1 2707 94 + 1 CACUACCAGCAGGCGGCCGAGUACCACGGCGGACCUCCCCCGCCA------CCGCAUCAUUACGGCGGUCCGCCGCCCCAGGGCCACUACGGAC------CGCCGC ........((.(((((((((((.....((((((((......(((.------............)))))))))))(((....))).))).))).)------)))))) ( -43.12) >DroYak_CAF1 286 94 + 1 CACUACCAGCAGGCGGCCGAGUAUCACGGCGGACCGCCCCCGCCA------CCGCAUCACUACGGCGGUCCGCCGCCCCAGGGCCACUAUGGAC------CGCCGC ........((.(((((((((((.....(((((((((((.......------............)))))))))))(((....))).))).))).)------)))))) ( -46.81) >DroMoj_CAF1 297 103 + 1 CACUACCAGCAGGCGGCCGAGUAUCAUGGCGGUCCGCCACCGC---CGCCGCCUCAUCACUACGGCGGACCGCCGCCCCAGGGGCAUUACGGUCCAUUGCCGCCGC ........((.((((((..........((((((.....)))))---)((((...........))))((((((..((((...))))....))))))...)))))))) ( -47.10) >consensus CACUACCAGCAGGCGGCCGAGUAUCACGGCGGACCGCCACCGCCA______CCGCAUCACUACGGCGGACCGCCGCCCCAGGGCCACUACGGAC______CGCCGC ............(((((.........((((((((((((.........................))))))))))))((..((.....))..)).........))))) (-31.52 = -31.11 + -0.41)

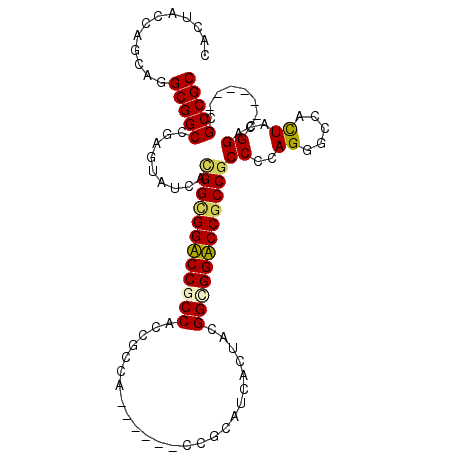
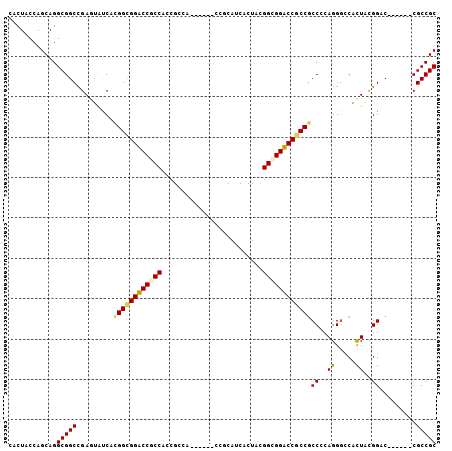
| Location | 19,706,035 – 19,706,129 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -51.28 |
| Consensus MFE | -33.56 |
| Energy contribution | -33.56 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19706035 94 - 22224390 GCGGCG------GUCCAUAGUGGCCCUGCGGUGGCGGACCGCCAUAGUGAUGCGG------UGGCGGUGGUGGUCCGCCGUGGUAUUCGGCCGCCUGCUGGUAGUG .(((((------(......((((((.(((.(((((((((((((((.((.((...)------).)).))))))))))))))).)))...))))))))))))...... ( -50.60) >DroVir_CAF1 297 94 - 1 GCGGCG------GACCGUAAUGGGCCUGCGGCGGCGGUCCACCAUAAUGAUGAGGCG------GCGGUGGCGGACCACCAUGAUACUCUGCCGCCUGCUGGUAAUG .(((((------((((((.....(((...))).)))))))............(((((------(((((((....)))))..(.....).)))))))))))...... ( -42.70) >DroGri_CAF1 306 100 - 1 GCGGCG------GGCCAUAGUGUCCCUGUGGCGGUGGUCCGCCAUAGUGAUGGGGCGGCGGUGGCGGCGGCGGACCGCCAUGAUACUCGGCCGCCUGCUGAUAAUG ((((((------((((((((.....)))))))(((((((((((.((....))..((.((....)).)))))))))))))...........))))).))........ ( -54.30) >DroSec_CAF1 2707 94 - 1 GCGGCG------GUCCGUAGUGGCCCUGGGGCGGCGGACCGCCGUAAUGAUGCGG------UGGCGGGGGAGGUCCGCCGUGGUACUCGGCCGCCUGCUGGUAGUG ((((((------((((((....(((....))).))))))))))))......((((------.((((((.....))))))(((((.....)))))))))........ ( -51.90) >DroYak_CAF1 286 94 - 1 GCGGCG------GUCCAUAGUGGCCCUGGGGCGGCGGACCGCCGUAGUGAUGCGG------UGGCGGGGGCGGUCCGCCGUGAUACUCGGCCGCCUGCUGGUAGUG ((((((------(((...(((((((....)))((((((((((((((....)))))------).((....))))))))))....)))).))))))).))........ ( -53.20) >DroMoj_CAF1 297 103 - 1 GCGGCGGCAAUGGACCGUAAUGCCCCUGGGGCGGCGGUCCGCCGUAGUGAUGAGGCGGCG---GCGGUGGCGGACCGCCAUGAUACUCGGCCGCCUGCUGGUAGUG ((((((((...(((((((...((((...)))).)))))))(((((.........)))))(---(((((.....))))))..........)))))).))........ ( -55.00) >consensus GCGGCG______GUCCAUAGUGGCCCUGGGGCGGCGGACCGCCAUAGUGAUGCGG______UGGCGGUGGCGGACCGCCAUGAUACUCGGCCGCCUGCUGGUAGUG (((((.......(.((.(((.....))).)))(((((((((((.........................)))))))))))..........)))))............ (-33.56 = -33.56 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:46 2006