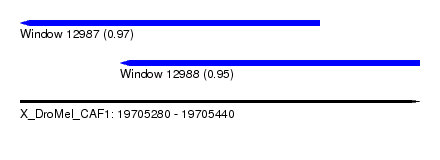
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,705,280 – 19,705,440 |
| Length | 160 |
| Max. P | 0.974994 |

| Location | 19,705,280 – 19,705,400 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -58.67 |
| Consensus MFE | -55.79 |
| Energy contribution | -55.13 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19705280 120 - 22224390 GUACUGCAUCUUGCAGUAGGCCUCCUGGGCGGCGCCAUUGCUGGCGUUGACGUAGACGCCGGUGAUGGUGACGUCCGUGUCCUGGACAAUGACGACCGACGAGGAGGUGGGCCGGCUGCA ...........((((((.((((((((.(.(((((((((..((((((((......))))))))..)))))).((((..((((...))))..)))).))).).))))....)))).)))))) ( -62.00) >DroEre_CAF1 12197 120 - 1 GUACUGCAUCUUGCAGUAGGCCUCCUGGGCGGCUCCGUUGCUGGCGUUCACGUAGACGCCGGUGAUGGUGACGUCCGUGUCCUGGACAAUGACCACCGACGAGGAGGUGGGCCGGCUGCA ...........((((((.(((((((((((((.(.((((..((((((((......))))))))..)))).).))))).((((.(((.......)))..))))...))).))))).)))))) ( -58.00) >DroYak_CAF1 8911 120 - 1 GUACUGCAUUUUGCAGUAGGCCUCCUGGGCGGCUCCGUUGCUGGCGUUCACAUAGACGCCGGUAAUGGUGACGUCCGUGUCCUGAACAAUGACGACUGACGAGGAGGUGGGCCGGCUGCA ...........((((((.(((((((((((((.(.((((((((((((((......)))))))))))))).).)))))(((((.(.....).))).))........))).))))).)))))) ( -56.00) >consensus GUACUGCAUCUUGCAGUAGGCCUCCUGGGCGGCUCCGUUGCUGGCGUUCACGUAGACGCCGGUGAUGGUGACGUCCGUGUCCUGGACAAUGACGACCGACGAGGAGGUGGGCCGGCUGCA ...........((((((.(((((((((((((.(.((((((((((((((......)))))))))))))).).)))))..(((.(.....).)))..((.....))))).))))).)))))) (-55.79 = -55.13 + -0.66)


| Location | 19,705,320 – 19,705,440 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -60.07 |
| Consensus MFE | -58.27 |
| Energy contribution | -57.83 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.950937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19705320 120 - 22224390 GGCGCAGGACGCGCUCCUCGGUGACGCUCGACUGGACGCGGUACUGCAUCUUGCAGUAGGCCUCCUGGGCGGCGCCAUUGCUGGCGUUGACGUAGACGCCGGUGAUGGUGACGUCCGUGU .((((((((((((.(((((((......))))..))))))).((((((.....))))))....))))(((((.((((((..((((((((......))))))))..)))))).))))))))) ( -64.10) >DroEre_CAF1 12237 120 - 1 GUCGCAGGACGCGCUCCUCGGUGACGCUCGACUGGACGCGGUACUGCAUCUUGCAGUAGGCCUCCUGGGCGGCUCCGUUGCUGGCGUUCACGUAGACGCCGGUGAUGGUGACGUCCGUGU ..(((((((((((.(((((((......))))..))))))).((((((.....))))))....))))(((((.(.((((..((((((((......))))))))..)))).).)))))))). ( -57.80) >DroYak_CAF1 8951 120 - 1 GUCGCAGGACGCGCUCCUCGGUGACGCUCGACUGGACGCGGUACUGCAUUUUGCAGUAGGCCUCCUGGGCGGCUCCGUUGCUGGCGUUCACAUAGACGCCGGUAAUGGUGACGUCCGUGU ..(((((((((((.(((((((......))))..))))))).((((((.....))))))....))))(((((.(.((((((((((((((......)))))))))))))).).)))))))). ( -58.30) >consensus GUCGCAGGACGCGCUCCUCGGUGACGCUCGACUGGACGCGGUACUGCAUCUUGCAGUAGGCCUCCUGGGCGGCUCCGUUGCUGGCGUUCACGUAGACGCCGGUGAUGGUGACGUCCGUGU ..(((((((((((.(((((((......))))..))))))).((((((.....))))))....))))(((((.(.((((((((((((((......)))))))))))))).).)))))))). (-58.27 = -57.83 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:44 2006