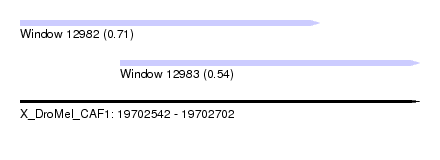
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,702,542 – 19,702,702 |
| Length | 160 |
| Max. P | 0.707946 |

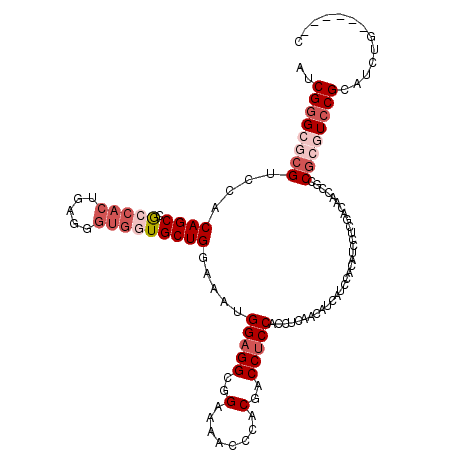
| Location | 19,702,542 – 19,702,662 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.75 |
| Mean single sequence MFE | -44.80 |
| Consensus MFE | -31.66 |
| Energy contribution | -33.22 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19702542 120 + 22224390 AUUCGGCGGCCAAGAAGCCGGCCAAGGAUCCAACGGGAUCAUCGGGCGUGUCCACAGCCACCACUGAGGGUGGUGCUGGAAAUGGAGGCGGAAAACAAACGACCUCCACCACGACAUCAU ...((((.........))))(((...(((((....)))))....)))(((((..((((.(((((.....)))))))))....((((((((.........)).))))))....)))))... ( -48.90) >DroSec_CAF1 8990 120 + 1 AUUCGGCGGCCAAGAAGCCGGCCAAGGAUCCCACGGGUACAUCGAGCGCGUCAACAGCCGCCACUGAGAAUGAUGCUGAACAUGGAGGAGGAAAACCCACGACCUCCACCUCCACGUCAU ....(((((((........))))..((.....(((.((.......)).)))......))))).......((((((.......((((((.(((............))).)))))))))))) ( -35.31) >DroSim_CAF1 8843 120 + 1 AUUCGGCGGCCAAGAAGCCGGCCAAGGAUCCCACGGGAUCAUCGGGCGCGUCCACAGCCACCACGGAGGGUGGUGCUGGAAAUGGAGGAGGAAAACCCACGACCUCCACCUCCACAUCAU ....((((((......))))(((...(((((....)))))....)))....)).((((.(((((.....)))))))))((..((((((.(((............))).))))))..)).. ( -49.20) >DroEre_CAF1 9522 117 + 1 AUUCGGCGGCCAAGAAGCCGGCCAAGGAUCCCACAGGAUCGUCGGGAUCGCUCACAGCCGCCACCGAGGG---UGCUGGAAACGGAGGCGGAAAACCCACAACCUCCACCUCAACAUCCU ....((((((......)))(((..((((((((((......)).)))))).))....))))))...((((.---....(....)(((((..(........)..))))).))))........ ( -43.90) >DroYak_CAF1 6287 120 + 1 ACACGGCGGCCAAGAAGCCGGCCAAGGAUCCCACAGGAUCAUCGGGAUCGGUCACAGCCGCCACUGAGGGUGGUGCUGGAAAUGGUGGCGGGAAACCAACGACCUCCACCUUAACAUCAA ....((((((......(((((((...(((((....)))))....)).)))))....))))))....(((((((..........(((.(.((....))..).))).)))))))........ ( -46.70) >consensus AUUCGGCGGCCAAGAAGCCGGCCAAGGAUCCCACGGGAUCAUCGGGCGCGUCCACAGCCGCCACUGAGGGUGGUGCUGGAAAUGGAGGCGGAAAACCCACGACCUCCACCUCAACAUCAU ....((((((......))).)))...(((((....)))))...((.........((((.(((((.....))))))))).....(((((..(........)..))))).)).......... (-31.66 = -33.22 + 1.56)



| Location | 19,702,582 – 19,702,702 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.86 |
| Mean single sequence MFE | -39.67 |
| Consensus MFE | -23.92 |
| Energy contribution | -26.08 |
| Covariance contribution | 2.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19702582 120 + 22224390 AUCGGGCGUGUCCACAGCCACCACUGAGGGUGGUGCUGGAAAUGGAGGCGGAAAACAAACGACCUCCACCACGACAUCAUCCACAUCCUCGACAACCGCCCCGUCCGCAUCUGCAUCUGC ...(((((((((..(((......))).((((((((((((...((((((((.........)).))))))))).).))))))))........))))..))))).....((....))...... ( -42.50) >DroSec_CAF1 9030 114 + 1 AUCGAGCGCGUCAACAGCCGCCACUGAGAAUGAUGCUGAACAUGGAGGAGGAAAACCCACGACCUCCACCUCCACGUCAUCCACAUCCUCGACAACCGACGCGUCCGCAUCUG------C ..((..((((((..(((......))).((..((((..((((.((((((.(((............))).)))))).))..))..)))).)).......))))))..))......------. ( -34.30) >DroSim_CAF1 8883 114 + 1 AUCGGGCGCGUCCACAGCCACCACGGAGGGUGGUGCUGGAAAUGGAGGAGGAAAACCCACGACCUCCACCUCCACAUCAUCCACAUCCUCGGCAACCGCCGCGUCCGCAUCUG------C ..(((((((...............(((((((((((.((((..((((((.((.....))....))))))..))))))))))))...)))..(((....))))))))))......------. ( -47.70) >DroEre_CAF1 9562 117 + 1 GUCGGGAUCGCUCACAGCCGCCACCGAGGG---UGCUGGAAACGGAGGCGGAAAACCCACAACCUCCACCUCAACAUCCUCCACUUCCUCGACAUCCGCCGCGUCCGCGUCCGCGUCCUC (.(((((.(((..((.((.((...((((((---((..(((....((((.(((............))).))))....)))..)))..)))))......)).))))..)))))).)).)... ( -37.40) >DroYak_CAF1 6327 112 + 1 AUCGGGAUCGGUCACAGCCGCCACUGAGGGUGGUGCUGGAAAUGGUGGCGGGAAACCAACGACCUCCACCUUAACAUCAACCACGUGCUCGACAUCCGCCGCAUCCGCUUCC-------- ...((((.(((((.((((.(((((.....)))))))))))....(((((((....))..(((....(((...............))).)))......)))))..))).))))-------- ( -36.46) >consensus AUCGGGCGCGUCCACAGCCGCCACUGAGGGUGGUGCUGGAAAUGGAGGCGGAAAACCCACGACCUCCACCUCAACAUCAUCCACAUCCUCGACAACCGCCGCGUCCGCAUCUG______C ..((((((((....((((.(((((.....))))))))).....(((((..(........)..)))))................................))))))))............. (-23.92 = -26.08 + 2.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:39 2006