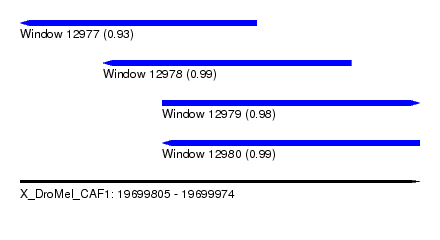
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,699,805 – 19,699,974 |
| Length | 169 |
| Max. P | 0.993437 |

| Location | 19,699,805 – 19,699,905 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 79.09 |
| Mean single sequence MFE | -21.73 |
| Consensus MFE | -19.01 |
| Energy contribution | -18.57 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19699805 100 - 22224390 GGAACUUAGUUCCACAAAGUAAAGUAAGUAAAUAAGUAAAGGUGGCAACUAAUGUCGUAAACAAUAUUACCGCUUGGGAAUCGAUUCUUUUCGAUUCUAA (((((...)))))..........................((((((....((((((.(....).)))))))))))).((((((((......)))))))).. ( -22.00) >DroSec_CAF1 6295 86 - 1 GUAACGUAGUUCCACAAAGUGGAG-------------AAAGGUGGAAGCUGAAACCGUAAGCGAUAUUACCGUUUGGGAAUCGAUUCCUUUCGAUUCUA- ..(((....((((((...))))))-------------...(((((..(((.........)))....))))))))..((((((((......)))))))).- ( -22.50) >DroSim_CAF1 6150 87 - 1 GGAACGUAGUUCCACAAAGUGGAG-------------AAAUGUGGAAGCUAAAGCCGUAAACGAUAUUACCGUUUGGUAAUCGAUUCCUUUCGAUUCUAA ((....((((((((((........-------------...))))).)))))...))..(((((.......)))))((.((((((......)))))))).. ( -20.70) >consensus GGAACGUAGUUCCACAAAGUGGAG_____________AAAGGUGGAAGCUAAAGCCGUAAACGAUAUUACCGUUUGGGAAUCGAUUCCUUUCGAUUCUAA ......(((((((((..........................)))).)))))((((.((((......)))).)))).((((((((......)))))))).. (-19.01 = -18.57 + -0.44)

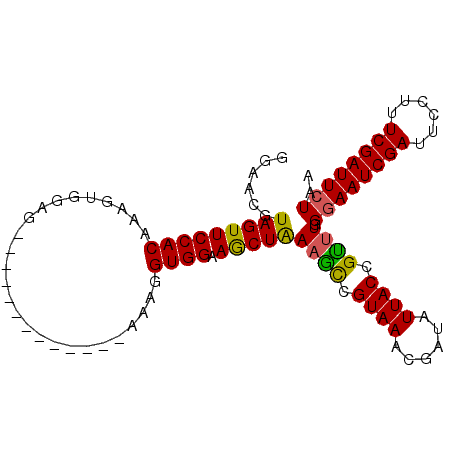

| Location | 19,699,840 – 19,699,945 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.47 |
| Mean single sequence MFE | -28.01 |
| Consensus MFE | -21.84 |
| Energy contribution | -21.07 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19699840 105 - 22224390 CAAGUACUUGCUAAGCGUGAGUGUUUGGCAUCCCAGAUGUGGAACUUAGUUCCACAAAGUAAAGUAAGUAAAUAAGUAAAGGUGGCAACUAAUGUCGUAAACAAU ....((((((((..((....))((((((....))))))(((((((...))))))).......)))))))).............((((.....))))......... ( -26.50) >DroSec_CAF1 6329 92 - 1 CAAGUACUUGCUAAGCGCGAGUGCUUGGCAUCACAGAAGUGUAACGUAGUUCCACAAAGUGGAG-------------AAAGGUGGAAGCUGAAACCGUAAGCGAU (((((((((((.....)))))))))))((..(((....)))..((((((((((((.........-------------....)))).)))))....)))..))... ( -27.42) >DroSim_CAF1 6185 92 - 1 CAAGUACUUGCUAAGCGCGAGUGCUUGGCAGCACAGAUGUGGAACGUAGUUCCACAAAGUGGAG-------------AAAUGUGGAAGCUAAAGCCGUAAACGAU .((((((((((.....))))))))))(((..(((...((((((((...))))))))..)))...-------------....((....))....)))......... ( -30.10) >consensus CAAGUACUUGCUAAGCGCGAGUGCUUGGCAUCACAGAUGUGGAACGUAGUUCCACAAAGUGGAG_____________AAAGGUGGAAGCUAAAGCCGUAAACGAU (((((((((((.....)))))))))))...........((...((((((((((((..........................)))).)))))....)))..))... (-21.84 = -21.07 + -0.77)

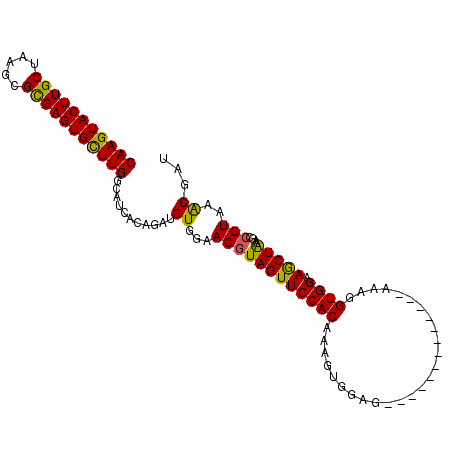

| Location | 19,699,865 – 19,699,974 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.91 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -11.69 |
| Energy contribution | -12.38 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19699865 109 + 22224390 UUUACUUAUUUACUUACUUUACUUUGUGGAACUAAGUUCCACAUCUGGGAUGCCAAACACUCACGCUUAGCAAGUACUUGCUCCUCUGCUAACCCA---------GUGCG-CCCAUA-AA ........................((((((((...))))))))..((((.(((...............(((((....)))))...(((......))---------).)))-))))..-.. ( -22.50) >DroSec_CAF1 6354 96 + 1 UUU-------------CUCCACUUUGUGGAACUACGUUACACUUCUGUGAUGCCAAGCACUCGCGCUUAGCAAGUACUUGCUCCUUUGCUAACCAA---------GUGCG-CCCAUA-AA ...-------------.......((((((.....(((((((....)))))))....(((((.(.(.((((((((..........)))))))))).)---------)))).-.)))))-). ( -24.50) >DroSim_CAF1 6210 96 + 1 UUU-------------CUCCACUUUGUGGAACUACGUUCCACAUCUGUGCUGCCAAGCACUCGCGCUUAGCAAGUACUUGCUCCUUUGCUAACCAA---------GUGCG-CCCAUA-AA ...-------------........((((((((...))))))))...(((((....))))).(((((((((((((..........)))))))....)---------)))))-......-.. ( -28.50) >DroYak_CAF1 3964 103 + 1 -----------------UCCACAAAGUGGACCUAUGUUCCACACCAGUGAUGCCAGAGUUUUCCACUUAGCGAGUACACACUCCCCUGCUAUCAAAUACGUAGCGAGGUAACUCAUAUAA -----------------..(((...(((((.......)))))....)))......(((((..((.......((((....))))...((((((.......)))))).)).)))))...... ( -22.10) >consensus UUU_____________CUCCACUUUGUGGAACUACGUUCCACAUCUGUGAUGCCAAACACUCGCGCUUAGCAAGUACUUGCUCCUCUGCUAACCAA_________GUGCG_CCCAUA_AA ...................(((..((((((((...))))))))...)))..................((((((((....)))....)))))............................. (-11.69 = -12.38 + 0.69)

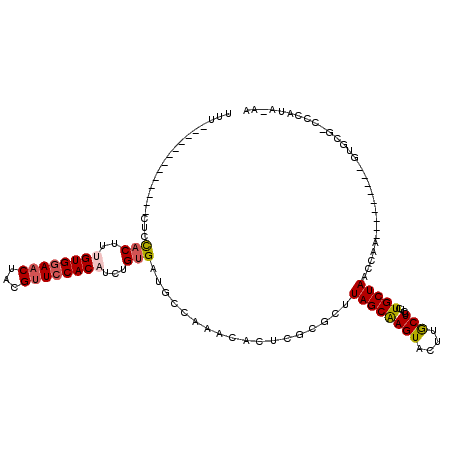

| Location | 19,699,865 – 19,699,974 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.91 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -16.98 |
| Energy contribution | -18.10 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19699865 109 - 22224390 UU-UAUGGG-CGCAC---------UGGGUUAGCAGAGGAGCAAGUACUUGCUAAGCGUGAGUGUUUGGCAUCCCAGAUGUGGAACUUAGUUCCACAAAGUAAAGUAAGUAAAUAAGUAAA ((-(((..(-(...(---------((((...((......)).......(((((((((....))))))))).))))).((((((((...)))))))).......))..)))))........ ( -32.80) >DroSec_CAF1 6354 96 - 1 UU-UAUGGG-CGCAC---------UUGGUUAGCAAAGGAGCAAGUACUUGCUAAGCGCGAGUGCUUGGCAUCACAGAAGUGUAACGUAGUUCCACAAAGUGGAG-------------AAA .(-((((..-.((((---------((.((..((.......(((((((((((.....)))))))))))))...))..))))))..)))))((((((...))))))-------------... ( -29.51) >DroSim_CAF1 6210 96 - 1 UU-UAUGGG-CGCAC---------UUGGUUAGCAAAGGAGCAAGUACUUGCUAAGCGCGAGUGCUUGGCAGCACAGAUGUGGAACGUAGUUCCACAAAGUGGAG-------------AAA ..-......-..(((---------((.....((....(..(((((((((((.....))))))))))).).)).....((((((((...)))))))))))))...-------------... ( -31.90) >DroYak_CAF1 3964 103 - 1 UUAUAUGAGUUACCUCGCUACGUAUUUGAUAGCAGGGGAGUGUGUACUCGCUAAGUGGAAAACUCUGGCAUCACUGGUGUGGAACAUAGGUCCACUUUGUGGA----------------- .((((((.....((..((((((....)).))))..))...)))))).((((.(((((((...((.((...((((....))))..)).)).))))))).)))).----------------- ( -29.00) >consensus UU_UAUGGG_CGCAC_________UUGGUUAGCAAAGGAGCAAGUACUUGCUAAGCGCGAGUGCUUGGCAUCACAGAUGUGGAACGUAGUUCCACAAAGUGGAG_____________AAA ...............................((......))...(((((((((((((....)))))))).........(((((((...))))))).)))))................... (-16.98 = -18.10 + 1.12)

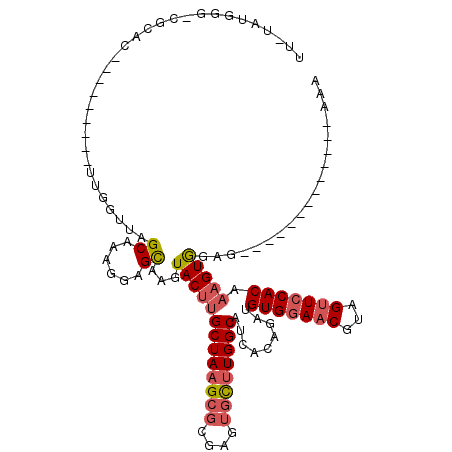
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:36 2006