| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,696,641 – 19,696,784 |
| Length | 143 |
| Max. P | 0.949801 |

| Location | 19,696,641 – 19,696,759 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 87.63 |
| Mean single sequence MFE | -54.37 |
| Consensus MFE | -42.46 |
| Energy contribution | -43.63 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
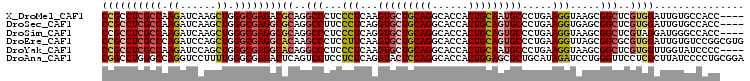
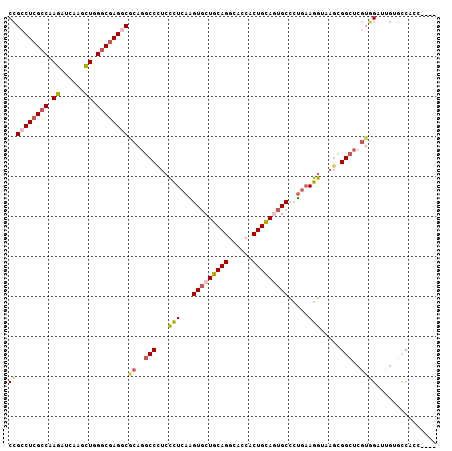
>X_DroMel_CAF1 19696641 118 + 22224390 CGGCUGGCAAAAAGCAGGCCGUACAAAGGGUUCAGCUUGGCCGCCUCGCCAAGAUCAAGCUGGGCGAGACGCAGGCCCUCCCUCAAGUGCUGCAGGCACCACUGCAAUGCCCUGAAGG (((((.((.....)).))))).....(((((...((..((((((((((((.((......)).))))))..)).)))).........((((.....))))....))...)))))..... ( -45.10) >DroSec_CAF1 3122 118 + 1 CGGCUGGCAAAAAGCAGGCCAUACAAAGGGUUCAGCUUGGCCGCCUCGCCAAGAUCAAGCUGGGCGAGGCGCAGGCCUUCCCUCAGGUGCUGCAGGCACCACUGCAGUGCCCUGAAGG .(((((((...((((.((((........))))..)))).)))((((((((.((......)).))))))))...))))..((.(((((..((((((......))))))..).)))).)) ( -60.30) >DroSim_CAF1 2292 118 + 1 CGGCUGGCAAAAAGCAGGCCAUACAAAGGGUUCAGCUUGGCCGCCUCGCCAAGAUCAAGCUGGGCGAGGCGCAGGCCCUCCCUCAGGUGCUGCAGGCACCACUGCAGUGCCCUGAAGG .((((.((.....)).)))).......(((....(((((..(((((((((.((......)).))))))))))))))...)))(((((..((((((......))))))..).))))... ( -60.20) >DroEre_CAF1 3392 118 + 1 CGGCUGGCAAAAAGCAGGCCGUACAAAGGGUUCAGCUUGGCCGCCUCGCCCAGAUCCAGCUGGGCGAGGCACAAGCCCUCCUUCAAGUGCUGCAGGCACCACUGCAGUGCCCUGAAGG .(((((((...((((.((((........))))..)))).)))(((((((((((......)))))))))))...))))..((((((.(..((((((......))))))..)..)))))) ( -66.60) >DroYak_CAF1 910 118 + 1 CGGCUGGCAAAAAGCAGGCCGUACAAAGGAUUCAGCUUGGCCGCCUCGCCAAGAUCCAGCUGGGCGAGGCACAGGCCCUCCCUCAAGUGCUGCAGGCACCACUGCAAUGCCCUGAAGG .(((((((...((((...((.......)).....)))).)))((((((((.((......)).))))))))...))))...(((..((.(((((((......)))))..)).))..))) ( -48.80) >DroAna_CAF1 14703 118 + 1 CGGCUGGCGAAGAGCAGCCCGUAAAGUGGGUUUAGCUUGGCGGCCUGGGCCAGGUCCUUUUGGGCGAGACUCAGUCCUCCUCUCAGGUACUCCAGGCACCACUGGAGCGCUGCAUAGA ...(((((...((((((((((.....))))))..))))(((((((((((....((((....))))(((........)))..))))))).((((((......)))))))))))).))). ( -45.20) >consensus CGGCUGGCAAAAAGCAGGCCGUACAAAGGGUUCAGCUUGGCCGCCUCGCCAAGAUCAAGCUGGGCGAGGCGCAGGCCCUCCCUCAAGUGCUGCAGGCACCACUGCAGUGCCCUGAAGG .((((.((.....)).))))......((((....(((((..(((((((((.((......)).))))))))))))))...))))...(((((((((......)))))))))........ (-42.46 = -43.63 + 1.17)

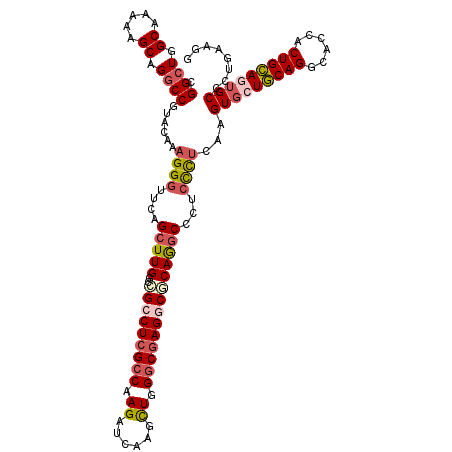

| Location | 19,696,641 – 19,696,759 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 87.63 |
| Mean single sequence MFE | -53.25 |
| Consensus MFE | -45.15 |
| Energy contribution | -45.05 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19696641 118 - 22224390 CCUUCAGGGCAUUGCAGUGGUGCCUGCAGCACUUGAGGGAGGGCCUGCGUCUCGCCCAGCUUGAUCUUGGCGAGGCGGCCAAGCUGAACCCUUUGUACGGCCUGCUUUUUGCCAGCCG ((((((((((..(((((......))))))).))))))))..(((..((((((((((.((......)).))))))))((((..((..........))..))))........))..))). ( -49.70) >DroSec_CAF1 3122 118 - 1 CCUUCAGGGCACUGCAGUGGUGCCUGCAGCACCUGAGGGAAGGCCUGCGCCUCGCCCAGCUUGAUCUUGGCGAGGCGGCCAAGCUGAACCCUUUGUAUGGCCUGCUUUUUGCCAGCCG ((((((((...((((((......))))))..))))))))..(((..((((((((((.((......)).))))))))(((((.((..........)).)))))........))..))). ( -59.40) >DroSim_CAF1 2292 118 - 1 CCUUCAGGGCACUGCAGUGGUGCCUGCAGCACCUGAGGGAGGGCCUGCGCCUCGCCCAGCUUGAUCUUGGCGAGGCGGCCAAGCUGAACCCUUUGUAUGGCCUGCUUUUUGCCAGCCG ((((((((...((((((......))))))..))))))))..(((..((((((((((.((......)).))))))))(((((.((..........)).)))))........))..))). ( -59.20) >DroEre_CAF1 3392 118 - 1 CCUUCAGGGCACUGCAGUGGUGCCUGCAGCACUUGAAGGAGGGCUUGUGCCUCGCCCAGCUGGAUCUGGGCGAGGCGGCCAAGCUGAACCCUUUGUACGGCCUGCUUUUUGCCAGCCG ((((((((...((((((......))))))..))))))))..((((((((((((((((((......))))))))))))..))))))............((((..((.....))..)))) ( -61.20) >DroYak_CAF1 910 118 - 1 CCUUCAGGGCAUUGCAGUGGUGCCUGCAGCACUUGAGGGAGGGCCUGUGCCUCGCCCAGCUGGAUCUUGGCGAGGCGGCCAAGCUGAAUCCUUUGUACGGCCUGCUUUUUGCCAGCCG ((((((((((..(((((......))))))).))))))))..((((...((((((((.((......)).)))))))))))).................((((..((.....))..)))) ( -52.30) >DroAna_CAF1 14703 118 - 1 UCUAUGCAGCGCUCCAGUGGUGCCUGGAGUACCUGAGAGGAGGACUGAGUCUCGCCCAAAAGGACCUGGCCCAGGCCGCCAAGCUAAACCCACUUUACGGGCUGCUCUUCGCCAGCCG .....((((((((((((......)))))))..(((((((..((....((..(.((((....))....(((....))))).)..))...))..)))).))))))))............. ( -37.70) >consensus CCUUCAGGGCACUGCAGUGGUGCCUGCAGCACCUGAGGGAGGGCCUGCGCCUCGCCCAGCUGGAUCUUGGCGAGGCGGCCAAGCUGAACCCUUUGUACGGCCUGCUUUUUGCCAGCCG ((((((((...((((((......))))))..))))))))..((((...((((((((.((......)).)))))))))))).................((((..((.....))..)))) (-45.15 = -45.05 + -0.10)

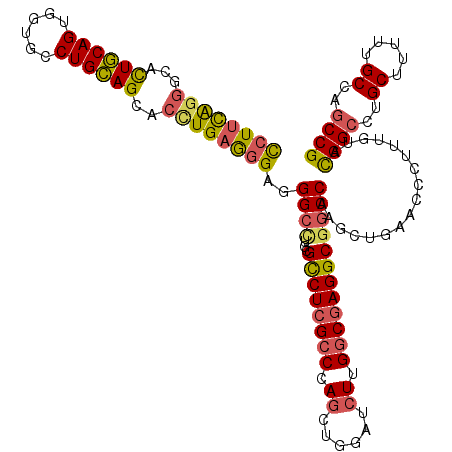

| Location | 19,696,681 – 19,696,784 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 79.06 |
| Mean single sequence MFE | -48.38 |
| Consensus MFE | -31.58 |
| Energy contribution | -32.20 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.949801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
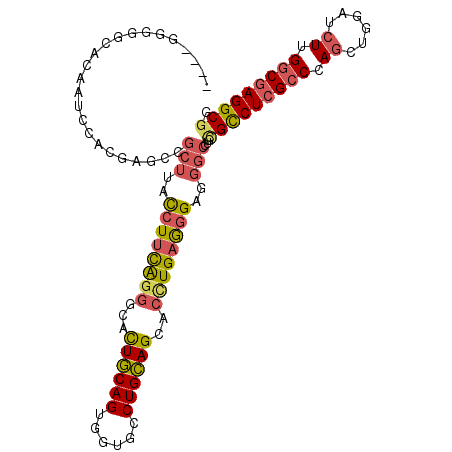
>X_DroMel_CAF1 19696681 103 + 22224390 CCGCCUCGCCAAGAUCAAGCUGGGCGAGACGCAGGCCCUCCCUCAAGUGCUGCAGGCACCACUGCAAUGCCCUGAAGGUAAGCGGCUCGUGGAUUGUGCCACC---- .((.((((((.((......)).)))))).))..((((((.(((..((.(((((((......)))))..)).))..)))..)).)))).((((......)))).---- ( -37.30) >DroSec_CAF1 3162 103 + 1 CCGCCUCGCCAAGAUCAAGCUGGGCGAGGCGCAGGCCUUCCCUCAGGUGCUGCAGGCACCACUGCAGUGCCCUGAAGGUGAGCGGCUCGUGGAUUGUGCCACC---- .(((((((((.((......)).)))))))))..((((((((((((((..((((((......))))))..).))).))).))).)))).((((......)))).---- ( -55.80) >DroSim_CAF1 2332 103 + 1 CCGCCUCGCCAAGAUCAAGCUGGGCGAGGCGCAGGCCCUCCCUCAGGUGCUGCAGGCACCACUGCAGUGCCCUGAAGGUAAGCGGCUCGUAGGAUGGGCCACC---- .(((((((((.((......)).)))))))))..((((((((.(((((..((((((......))))))..).))))......(((...))).))).)))))...---- ( -55.80) >DroEre_CAF1 3432 107 + 1 CCGCCUCGCCCAGAUCCAGCUGGGCGAGGCACAAGCCCUCCUUCAAGUGCUGCAGGCACCACUGCAGUGCCCUGAAGGUUAGCGGCGCGUGGAUUGUGUCCGGCGUG ..(((((((((((......)))))))))))....((...((((((.(..((((((......))))))..)..))))))...)).((((.(((((...))))))))). ( -61.20) >DroYak_CAF1 950 103 + 1 CCGCCUCGCCAAGAUCCAGCUGGGCGAGGCACAGGCCCUCCCUCAAGUGCUGCAGGCACCACUGCAAUGCCCUGAAGGUAAGCGGCUCGUGGUUGGUAUCCCC---- ((((((((((.((......)).))))))))((..(((((.(((..((.(((((((......)))))..)).))..)))..)).)))..))))...........---- ( -41.50) >DroAna_CAF1 14743 107 + 1 CGGCCUGGGCCAGGUCCUUUUGGGCGAGACUCAGUCCUCCUCUCAGGUACUCCAGGCACCACUGGAGCGCUGCAUAGAUCCUGGGUUCCUCGCUUAUCCCCUGCGGA (.((..((((((((....)))))(((((((((((...((......((..((((((......))))))..)).....))..))))))..))))).....))).)).). ( -38.70) >consensus CCGCCUCGCCAAGAUCAAGCUGGGCGAGGCGCAGGCCCUCCCUCAAGUGCUGCAGGCACCACUGCAGUGCCCUGAAGGUAAGCGGCUCGUGGAUUGUGCCACC____ ((((((((((.((......)).))))))))((..(((...(((...(((((((((......))))))))).....))).....)))..))))............... (-31.58 = -32.20 + 0.62)

| Location | 19,696,681 – 19,696,784 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 79.06 |
| Mean single sequence MFE | -48.82 |
| Consensus MFE | -36.97 |
| Energy contribution | -37.20 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19696681 103 - 22224390 ----GGUGGCACAAUCCACGAGCCGCUUACCUUCAGGGCAUUGCAGUGGUGCCUGCAGCACUUGAGGGAGGGCCUGCGUCUCGCCCAGCUUGAUCUUGGCGAGGCGG ----.((((......))))..((.(((..((((((((((..(((((......))))))).))))))))..)))..))((((((((.((......)).)))))))).. ( -46.50) >DroSec_CAF1 3162 103 - 1 ----GGUGGCACAAUCCACGAGCCGCUCACCUUCAGGGCACUGCAGUGGUGCCUGCAGCACCUGAGGGAAGGCCUGCGCCUCGCCCAGCUUGAUCUUGGCGAGGCGG ----.((((......))))..(((.....((((((((...((((((......))))))..))))))))..)))...(((((((((.((......)).))))))))). ( -51.90) >DroSim_CAF1 2332 103 - 1 ----GGUGGCCCAUCCUACGAGCCGCUUACCUUCAGGGCACUGCAGUGGUGCCUGCAGCACCUGAGGGAGGGCCUGCGCCUCGCCCAGCUUGAUCUUGGCGAGGCGG ----...(((((.((....))........((((((((...((((((......))))))..)))))))).)))))..(((((((((.((......)).))))))))). ( -54.10) >DroEre_CAF1 3432 107 - 1 CACGCCGGACACAAUCCACGCGCCGCUAACCUUCAGGGCACUGCAGUGGUGCCUGCAGCACUUGAAGGAGGGCUUGUGCCUCGCCCAGCUGGAUCUGGGCGAGGCGG ..(((.(((.....)))..)))(((((..((((((((...((((((......))))))..))))))))..)))....(((((((((((......))))))))))))) ( -55.80) >DroYak_CAF1 950 103 - 1 ----GGGGAUACCAACCACGAGCCGCUUACCUUCAGGGCAUUGCAGUGGUGCCUGCAGCACUUGAGGGAGGGCCUGUGCCUCGCCCAGCUGGAUCUUGGCGAGGCGG ----.((....))..(((((.(((.((..((((((((((..(((((......))))))).))))))))))))).)))((((((((.((......)).)))))))))) ( -47.20) >DroAna_CAF1 14743 107 - 1 UCCGCAGGGGAUAAGCGAGGAACCCAGGAUCUAUGCAGCGCUCCAGUGGUGCCUGGAGUACCUGAGAGGAGGACUGAGUCUCGCCCAAAAGGACCUGGCCCAGGCCG (((....((.....((((((....(((..(((...(((.(((((((......)))))))..)))...)))...)))..))))))))....)))...(((....))). ( -37.40) >consensus ____GGGGGCACAAUCCACGAGCCGCUUACCUUCAGGGCACUGCAGUGGUGCCUGCAGCACCUGAGGGAGGGCCUGCGCCUCGCCCAGCUGGAUCUUGGCGAGGCGG ........................(((..((((((((...((((((......))))))..))))))))..)))...(((((((((.((......)).))))))))). (-36.97 = -37.20 + 0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:32 2006