
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,686,336 – 19,686,438 |
| Length | 102 |
| Max. P | 0.622380 |

| Location | 19,686,336 – 19,686,438 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -40.68 |
| Consensus MFE | -22.98 |
| Energy contribution | -23.78 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19686336 102 - 22224390 AGCCACUGCGAUUGGACACCGCGGUCUGAAAAGUGUUGUACAAAUGCUUGUACGGCGUCCACAUGUCCUCGAUUACAGCUGUGGACG-------CAUGGGU-GGCA---------UUAG .((((((((..(..(((......)))..)...))..(((((((....)))))))((((((((((((........)))..))))))))-------)...)))-))).---------.... ( -37.90) >DroSec_CAF1 59947 102 - 1 AGCCACUGCCAUUGGACACCGACGCCUGAAAAACGCUGUACAGAUGCUUGUACGGCGUCCACAUGUCCUCGGUUAUACCUGUGGACG-------CAUGGGU-GGCA---------UUAG .(((((.((..((((...)))).)).......(((((((((((....)))))))))))(((..(((((.(((......))).)))))-------..)))))-))).---------.... ( -38.50) >DroSim_CAF1 47179 111 - 1 AGCUACUGCCAUUGGACACCGUCGUCUGAAAAACGCUGUACAGAUGCUUGUACGGCGUCCACAUGUCCUCGGUUAUGCCUGUGGACG-------CAUGGGU-GGCAUUAGUGGCAUUAG .(((((((((((..(((......)))..)...(((((((((((....)))))))))))(((..(((((.(((......))).)))))-------..))).)-)))...))))))..... ( -45.80) >DroEre_CAF1 71199 109 - 1 CGGCACAGGGAUUGGACACAGUCGCCUGGAAAACGCUAUACAGAUGCUUGUACGGCGUCCACAUGUCCUCGGUCACACCUGUGGACGG-GACGGAAUGGGCGGGCA---------UUAG ..((.(((((((((....))))).))))......((..(((((....)))))..))(((((..(((((.((..(((....)))..)))-))))...)))))..)).---------.... ( -43.30) >DroYak_CAF1 73329 99 - 1 CGCCACUGGGAUUGGACACCGUCGCCUGGAAAACGCUGUACAGAUGCUUGUACGGCGUCCACAUGUCCUCGGUCACACCUGUGGACG-----------GGCGGGCA---------UUAG .(((...((.........))..((((((((...((((((((((....)))))))))))))....((((.(((......))).)))))-----------))))))).---------.... ( -40.60) >DroAna_CAF1 86398 107 - 1 CGCCGGUGGGGUUCGAGACGGCCUGCUGGAAAACGCUGUAGAGAUGCUUGUACGGCGUCCACAUGUCCUCGGUUACACCUGCAAGUGGCGGCGGAGUGG---GGGA---------UGAG ((((((((....(((((..(((.((.((((...(((((((.((....)).))))))))))))).))))))))...)))).((.....))))))......---....---------.... ( -38.00) >consensus AGCCACUGCGAUUGGACACCGUCGCCUGAAAAACGCUGUACAGAUGCUUGUACGGCGUCCACAUGUCCUCGGUUACACCUGUGGACG_______CAUGGGU_GGCA_________UUAG .(((........(((((.........(....)..(((((((((....))))))))))))))...((((.(((......))).))))................))).............. (-22.98 = -23.78 + 0.81)

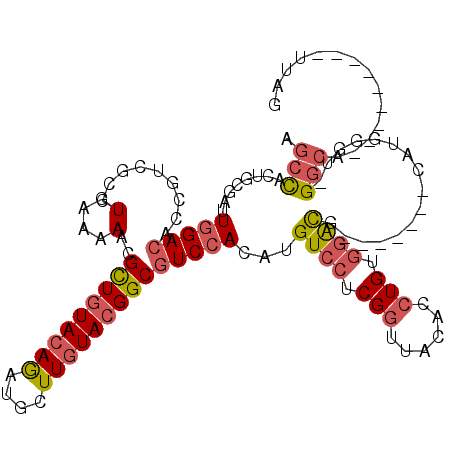
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:28 2006