| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,682,433 – 19,682,588 |
| Length | 155 |
| Max. P | 0.913115 |

| Location | 19,682,433 – 19,682,553 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -49.05 |
| Consensus MFE | -41.79 |
| Energy contribution | -42.23 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19682433 120 + 22224390 GCGCAGCAACUCGAACUCAAACACAACAGGCGCGAGCGAGAGCGCCGACAGCGCUGCCAACUGGACUGCAGAUGCAAGCGAGUUGGCAGCCCUGGAUGUCGGAUGCAAGUGCGUACACGG ((((((((.(.(((..((..........(((((........)))))..(((.((((((((((.(..(((....)))..).)))))))))).)))))..)))).)))...)))))...... ( -53.10) >DroSec_CAF1 56257 120 + 1 GCGCAGCAACUCGAACUCAAAUACAACAGGAGCGAGCGAGAGCGCCGACGGCGCUGCCAACCGGACUGCAGAUGCAAGCGAGUUGGCAGCUCUGGACGUCGGAUGCAAGUGCGUACACGG (((((....((((..(((...........)))....)))).(((((((((..(((((((((((...(((....)))..)).)))))))))......)))))).)))...)))))...... ( -47.40) >DroSim_CAF1 43203 120 + 1 GCGCAGCAACUGGAACUCAAACACAACAGGCGCGAGCGAGAGCGCCGACGGCGCUGCCAACCGGACUGCAGAUGCAAGCGAGUUGGCAGCUCUGGACGUCGGAUGCAAGUGCAUACACGG ((((.(((.((((...((..........(((((........)))))..(((.(((((((((((...(((....)))..)).))))))))).)))))..)))).)))..))))........ ( -49.30) >DroEre_CAF1 67452 120 + 1 GCGCAGCAACUCGAACUCAAACACAACAGGCGCGAGCGAGAGAGCCGACGGCGCUGCCAACCGGACUGCAGAUGCAGGCAAGUUGGCAGUACUGGACGUCGGAUGCAAACGCGUACACGG (((..(((.(((...(((...(......)((....))))).)))((((((..(((((((((..(.((((....)))).)..)))))))))......)))))).)))...)))........ ( -46.40) >consensus GCGCAGCAACUCGAACUCAAACACAACAGGCGCGAGCGAGAGCGCCGACGGCGCUGCCAACCGGACUGCAGAUGCAAGCGAGUUGGCAGCUCUGGACGUCGGAUGCAAGUGCGUACACGG ((((.(((.(.(((..((..........(((((........)))))..(((.(((((((((..(..(((....)))..)..))))))))).)))))..)))).)))..))))........ (-41.79 = -42.23 + 0.44)

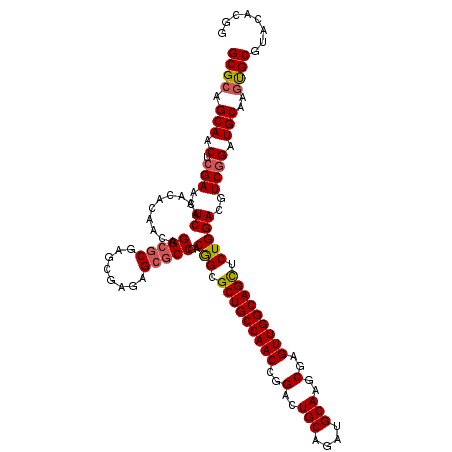

| Location | 19,682,433 – 19,682,553 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -48.88 |
| Consensus MFE | -44.49 |
| Energy contribution | -44.80 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19682433 120 - 22224390 CCGUGUACGCACUUGCAUCCGACAUCCAGGGCUGCCAACUCGCUUGCAUCUGCAGUCCAGUUGGCAGCGCUGUCGGCGCUCUCGCUCGCGCCUGUUGUGUUUGAGUUCGAGUUGCUGCGC ..(((((.(((((((..((.(((((.(((.((((((((((.(.((((....)))).).)))))))))).)))..(((((........)))))....))))).))...)))).)))))))) ( -54.60) >DroSec_CAF1 56257 120 - 1 CCGUGUACGCACUUGCAUCCGACGUCCAGAGCUGCCAACUCGCUUGCAUCUGCAGUCCGGUUGGCAGCGCCGUCGGCGCUCUCGCUCGCUCCUGUUGUAUUUGAGUUCGAGUUGCUGCGC ..(((((.(((...((..((((((......((((((((((.(.((((....)))).).))))))))))..)))))).)).((((...((((...........)))).)))).)))))))) ( -46.10) >DroSim_CAF1 43203 120 - 1 CCGUGUAUGCACUUGCAUCCGACGUCCAGAGCUGCCAACUCGCUUGCAUCUGCAGUCCGGUUGGCAGCGCCGUCGGCGCUCUCGCUCGCGCCUGUUGUGUUUGAGUUCCAGUUGCUGCGC ..(((((.(((((.((..((((((......((((((((((.(.((((....)))).).))))))))))..)))))).)).((((..((((.....))))..))))....)).)))))))) ( -46.60) >DroEre_CAF1 67452 120 - 1 CCGUGUACGCGUUUGCAUCCGACGUCCAGUACUGCCAACUUGCCUGCAUCUGCAGUCCGGUUGGCAGCGCCGUCGGCUCUCUCGCUCGCGCCUGUUGUGUUUGAGUUCGAGUUGCUGCGC ........((((..(((.((((((....(..(((((((((.(.((((....)))).).)))))))))..)))))))..(((..((((((((.....))))..))))..))).))).)))) ( -48.20) >consensus CCGUGUACGCACUUGCAUCCGACGUCCAGAGCUGCCAACUCGCUUGCAUCUGCAGUCCGGUUGGCAGCGCCGUCGGCGCUCUCGCUCGCGCCUGUUGUGUUUGAGUUCGAGUUGCUGCGC ..(((((.(((.......((((((....(.((((((((((.(.((((....)))).).)))))))))).))))))).((((..((((((((.....))))..))))..)))))))))))) (-44.49 = -44.80 + 0.31)

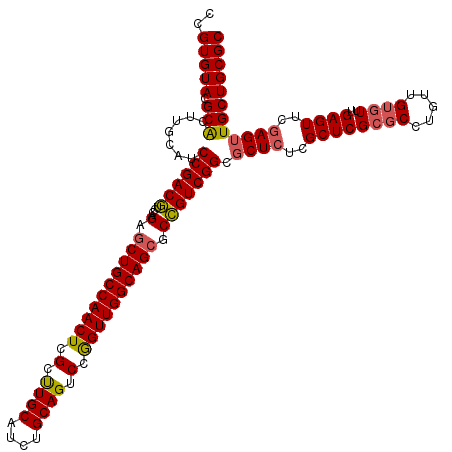

| Location | 19,682,473 – 19,682,588 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.80 |
| Mean single sequence MFE | -45.24 |
| Consensus MFE | -33.84 |
| Energy contribution | -33.60 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19682473 115 + 22224390 AGCGCCGACAGCGCUGCCAACUGGACUGCAGAUGCAAGCGAGUUGGCAGCCCUGGAUGUCGGAUGCAAGUGCGUACACGGUUAGAAAUCGGGGAUAGCA---AAACUAGUGCUGCCCG-- .(((((((((..((((((((((.(..(((....)))..).))))))))))......)))))).)))...........((((.....))))(((.(((((---.......)))))))).-- ( -49.40) >DroSec_CAF1 56297 118 + 1 AGCGCCGACGGCGCUGCCAACCGGACUGCAGAUGCAAGCGAGUUGGCAGCUCUGGACGUCGGAUGCAAGUGCGUACACGGUUAGAAAUCAGGGAUAGCAACGAAACUAGUGCUGCCCG-- .(((((((((..(((((((((((...(((....)))..)).)))))))))......)))))).)))..(((....)))............(((.(((((.(.......))))))))).-- ( -47.20) >DroSim_CAF1 43243 115 + 1 AGCGCCGACGGCGCUGCCAACCGGACUGCAGAUGCAAGCGAGUUGGCAGCUCUGGACGUCGGAUGCAAGUGCAUACACGGUUAGAAAUCGGGGAUAGCAACGAAACUAGUG---CCCG-- .(((((((((..(((((((((((...(((....)))..)).)))))))))......)))))).)))..(((....)))..........((((.((((........)).)).---))))-- ( -43.00) >DroEre_CAF1 67492 120 + 1 AGAGCCGACGGCGCUGCCAACCGGACUGCAGAUGCAGGCAAGUUGGCAGUACUGGACGUCGGAUGCAAACGCGUACACGGUUAGAAAUCGGGGAUUGCAACGGUACUAGUGCUUCUCGCA ..(((((.(((..((((((((..(.((((....)))).)..))))))))..))).(((.((........)))))...)))))......((((((..(((.(.......)))))))))).. ( -43.10) >DroYak_CAF1 69720 101 + 1 AGAGCCGACGGCGCUGCCAACCGGGCUGCAGAUGCAAGCAAGUUGGCAGCGCUGGACGACGGAUGCAAGUGCGUACACGGUUACAAAUCGGGGAUAACAAC------------------- ....(((((((((((((((((...(((((....)).)))..))))))))))))).....((.((((....))))...))........))))..........------------------- ( -43.50) >consensus AGCGCCGACGGCGCUGCCAACCGGACUGCAGAUGCAAGCGAGUUGGCAGCUCUGGACGUCGGAUGCAAGUGCGUACACGGUUAGAAAUCGGGGAUAGCAACGAAACUAGUGCU_CCCG__ ....(((((((.(((((((((..(..(((....)))..)..))))))))).)))..(((...((((....))))..)))........))))............................. (-33.84 = -33.60 + -0.24)

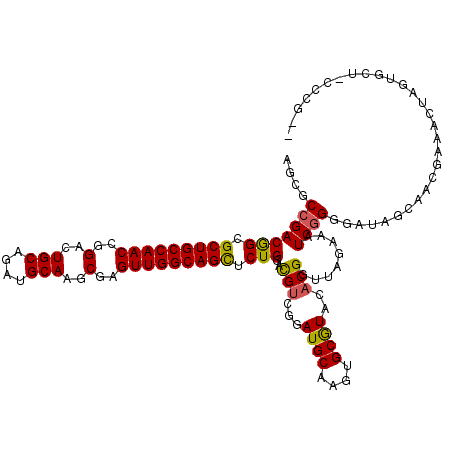
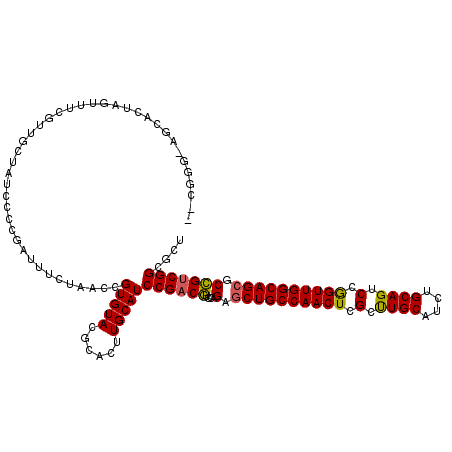
| Location | 19,682,473 – 19,682,588 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.80 |
| Mean single sequence MFE | -42.64 |
| Consensus MFE | -32.18 |
| Energy contribution | -31.94 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19682473 115 - 22224390 --CGGGCAGCACUAGUUU---UGCUAUCCCCGAUUUCUAACCGUGUACGCACUUGCAUCCGACAUCCAGGGCUGCCAACUCGCUUGCAUCUGCAGUCCAGUUGGCAGCGCUGUCGGCGCU --((((.((((.......---))))...))))..........(((....)))..((..((((((....(.((((((((((.(.((((....)))).).)))))))))).))))))).)). ( -45.10) >DroSec_CAF1 56297 118 - 1 --CGGGCAGCACUAGUUUCGUUGCUAUCCCUGAUUUCUAACCGUGUACGCACUUGCAUCCGACGUCCAGAGCUGCCAACUCGCUUGCAUCUGCAGUCCGGUUGGCAGCGCCGUCGGCGCU --.(((.(((((.......).))))..)))............(((....)))..((..((((((......((((((((((.(.((((....)))).).))))))))))..)))))).)). ( -42.60) >DroSim_CAF1 43243 115 - 1 --CGGG---CACUAGUUUCGUUGCUAUCCCCGAUUUCUAACCGUGUAUGCACUUGCAUCCGACGUCCAGAGCUGCCAACUCGCUUGCAUCUGCAGUCCGGUUGGCAGCGCCGUCGGCGCU --((((---...((((......))))..))))..........(((....)))..((..((((((......((((((((((.(.((((....)))).).))))))))))..)))))).)). ( -44.40) >DroEre_CAF1 67492 120 - 1 UGCGAGAAGCACUAGUACCGUUGCAAUCCCCGAUUUCUAACCGUGUACGCGUUUGCAUCCGACGUCCAGUACUGCCAACUUGCCUGCAUCUGCAGUCCGGUUGGCAGCGCCGUCGGCUCU ...(((..(((...(((((((((.((((...))))..))).)).)))).....)))..((((((....(..(((((((((.(.((((....)))).).)))))))))..)))))))))). ( -42.40) >DroYak_CAF1 69720 101 - 1 -------------------GUUGUUAUCCCCGAUUUGUAACCGUGUACGCACUUGCAUCCGUCGUCCAGCGCUGCCAACUUGCUUGCAUCUGCAGCCCGGUUGGCAGCGCCGUCGGCUCU -------------------..........(((((.(((((..(((....))))))))...........((((((((((((.(((.((....)))))..)))))))))))).))))).... ( -38.70) >consensus __CGGG_AGCACUAGUUUCGUUGCUAUCCCCGAUUUCUAACCGUGUACGCACUUGCAUCCGACGUCCAGAGCUGCCAACUCGCUUGCAUCUGCAGUCCGGUUGGCAGCGCCGUCGGCGCU ..........................................(((((......)))))((((((....(.((((((((((.(.((((....)))).).)))))))))).))))))).... (-32.18 = -31.94 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:25 2006