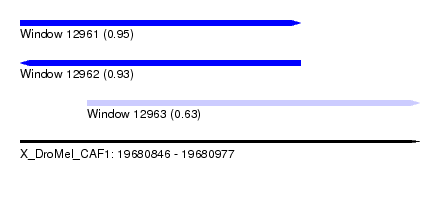
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,680,846 – 19,680,977 |
| Length | 131 |
| Max. P | 0.947754 |

| Location | 19,680,846 – 19,680,938 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.26 |
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -16.67 |
| Energy contribution | -17.12 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
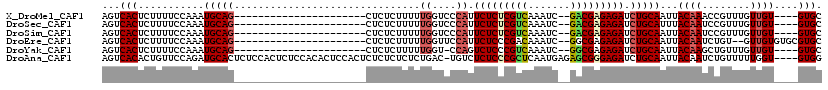
>X_DroMel_CAF1 19680846 92 + 22224390 AGUCACUCUUUUCCAAAUGCAG----------------------CUCUCUUUUUGGUCCCAUUCUCUCGUCAAAUC--GACGAGAGAUCUGCAAUUACAAACCGUUUGUUGU----GUGC ...(((...........(((((----------------------.........((....)).(((((((((.....--))))))))).)))))...((((........))))----))). ( -22.20) >DroSec_CAF1 54679 92 + 1 AGUCACUCUUUUCCAAAUGCAG----------------------CUCUCUUUUUGGUCCCAUUCUCUCGUCAAAUC--GACGAGAGAUCUGCAUUUACAAUCCGUUUGUUGU----GUGC ...(((........((((((((----------------------.........((....)).(((((((((.....--))))))))).))))))))(((((......)))))----))). ( -26.30) >DroSim_CAF1 41594 92 + 1 AGUCACUCUUUUCCAAAUGCAG----------------------CUCUCUUUUUGGUCCCAUUCUCUCGUCAAAUC--GACGAGAGAUCUGCAAUUACAAUCCGUUUGUUGU----GUGC ...(((...........(((((----------------------.........((....)).(((((((((.....--))))))))).)))))...(((((......)))))----))). ( -23.50) >DroEre_CAF1 65894 94 + 1 AGUCACUCUUUUCCAAAUGCAG----------------------CUCUCUUUUUGGUUCCAUUCUCCCGACAAAUC--GGCGAGAGAUCUGCAAUUACAAUCUGU--GUUGUGUGCGUGC ...(((...........(((((----------------------.((((((..((....)).....((((....))--)).)))))).)))))..((((((....--))))))...))). ( -21.80) >DroYak_CAF1 68149 91 + 1 AGUCACUCUUUUCCAAAUGCAG----------------------CUCUCUUUUUGGU-CCAGUCUCCCGUCAAAUC--GGCGAGAGAUCUGCAAUUACAAGCUGUUUGUUGU----GUGC .............(((..((((----------------------((......(((..-.(((((((.((((.....--)))).)))).)))))).....))))))...))).----.... ( -21.70) >DroAna_CAF1 79666 115 + 1 AGUCACACUGUUCCAGAUGCACUCUCCACUCUCCACACUCCACUCUCUCUCUCUGAC-UGUCUCUCCCGCUCAAUGAGAGCGGGAGAUCUGCAAUUACAAUCUGUUUUUGGU----GUGG ...(((((((...((((((((....................((..((.......)).-.)).((((((((((.....))))))))))..)))).......))))....))))----))). ( -32.81) >consensus AGUCACUCUUUUCCAAAUGCAG______________________CUCUCUUUUUGGUCCCAUUCUCCCGUCAAAUC__GACGAGAGAUCUGCAAUUACAAUCCGUUUGUUGU____GUGC ...(((...........(((((...............................((....)).(((((((((.......))))))))).)))))...((((........))))....))). (-16.67 = -17.12 + 0.45)
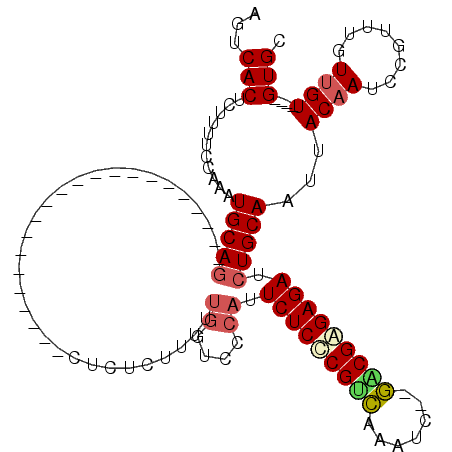
| Location | 19,680,846 – 19,680,938 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.26 |
| Mean single sequence MFE | -26.84 |
| Consensus MFE | -18.47 |
| Energy contribution | -18.63 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19680846 92 - 22224390 GCAC----ACAACAAACGGUUUGUAAUUGCAGAUCUCUCGUC--GAUUUGACGAGAGAAUGGGACCAAAAAGAGAG----------------------CUGCAUUUGGAAAAGAGUGACU ....----.......((..(((.(((.(((((.(((((((((--.....))))))))).((....)).........----------------------))))).))).)))...)).... ( -25.70) >DroSec_CAF1 54679 92 - 1 GCAC----ACAACAAACGGAUUGUAAAUGCAGAUCUCUCGUC--GAUUUGACGAGAGAAUGGGACCAAAAAGAGAG----------------------CUGCAUUUGGAAAAGAGUGACU ...(----((.((((.....))))((((((((.(((((((((--.....))))))))).((....)).........----------------------))))))))........)))... ( -27.80) >DroSim_CAF1 41594 92 - 1 GCAC----ACAACAAACGGAUUGUAAUUGCAGAUCUCUCGUC--GAUUUGACGAGAGAAUGGGACCAAAAAGAGAG----------------------CUGCAUUUGGAAAAGAGUGACU ...(----((.((((.....))))...(((((.(((((((((--.....))))))))).((....)).........----------------------)))))...........)))... ( -24.80) >DroEre_CAF1 65894 94 - 1 GCACGCACACAAC--ACAGAUUGUAAUUGCAGAUCUCUCGCC--GAUUUGUCGGGAGAAUGGAACCAAAAAGAGAG----------------------CUGCAUUUGGAAAAGAGUGACU ............(--((...((.(((.(((((..(((((.((--((....)))).....((....))....)))))----------------------))))).))).))....)))... ( -22.30) >DroYak_CAF1 68149 91 - 1 GCAC----ACAACAAACAGCUUGUAAUUGCAGAUCUCUCGCC--GAUUUGACGGGAGACUGG-ACCAAAAAGAGAG----------------------CUGCAUUUGGAAAAGAGUGACU ...(----((.((((.....))))...(((((..(((((...--..((((.(((....))).-..))))..)))))----------------------)))))...........)))... ( -22.30) >DroAna_CAF1 79666 115 - 1 CCAC----ACCAAAAACAGAUUGUAAUUGCAGAUCUCCCGCUCUCAUUGAGCGGGAGAGACA-GUCAGAGAGAGAGAGUGGAGUGUGGAGAGUGGAGAGUGCAUCUGGAACAGUGUGACU .(((----((......(((((.(((.(..(...((((((((((.....)))))))))).(((-.((..............)).))).....)..)....)))))))).....)))))... ( -38.14) >consensus GCAC____ACAACAAACAGAUUGUAAUUGCAGAUCUCUCGCC__GAUUUGACGAGAGAAUGGGACCAAAAAGAGAG______________________CUGCAUUUGGAAAAGAGUGACU ....................((.(((.(((((.(((((((((.......))))))))).((....))...............................))))).))).)).......... (-18.47 = -18.63 + 0.17)


| Location | 19,680,868 – 19,680,977 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 87.62 |
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -25.36 |
| Energy contribution | -25.52 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19680868 109 + 22224390 CUCUCUUUUUGGUCCCAUUCUCUCGUCAAAUCGACGAGAGAUCUGCAAUUACAAACCGUUUGUUGU----GUGCGUGCCAGUGGCGUAUGCAGUGAAUGGUAAAGGGGGGCUG ..........((((((..(((((((((.....))))))))).............((((((..((((----((((((.......))))))))))..)))))).....)))))). ( -46.10) >DroSec_CAF1 54701 104 + 1 CUCUCUUUUUGGUCCCAUUCUCUCGUCAAAUCGACGAGAGAUCUGCAUUUACAAUCCGUUUGUUGU----GUGCGUGCCAGUGGCGUAUGCAGUGAAUUGUAAGG-GG----G ............((((..(((((((((.....))))))))).......(((((....(((..((((----((((((.......))))))))))..))))))))))-))----. ( -36.80) >DroSim_CAF1 41616 105 + 1 CUCUCUUUUUGGUCCCAUUCUCUCGUCAAAUCGACGAGAGAUCUGCAAUUACAAUCCGUUUGUUGU----GUGCGUGCCAGUGGCGUAUGCAGUGAAUUGUGAGGGGG----G ............((((..(((((((((.....))))))))).......(((((....(((..((((----((((((.......))))))))))..)))))))))))).----. ( -37.00) >DroEre_CAF1 65916 106 + 1 CUCUCUUUUUGGUUCCAUUCUCCCGACAAAUCGGCGAGAGAUCUGCAAUUACAAUCUGU--GUUGUGUGCGUGCGUGCCAGUGGCGUAUGCAGAAAAUCGUAGGGGGG----- ((((((...(((((...(((((((((....)))).))))).((((((..((((((....--))))))(((((((......)).))))))))))).))))).)))))).----- ( -33.50) >DroYak_CAF1 68171 104 + 1 CUCUCUUUUUGGU-CCAGUCUCCCGUCAAAUCGGCGAGAGAUCUGCAAUUACAAGCUGUUUGUUGU----GUGCGUGCCAGUGGUGUACGUAGUAAAUCGUAAGGGGG----G ((((((((.((((-.(((((((.((((.....)))).)))).)))...((((.(((.....)))..----.((((((((....).))))))))))))))).)))))))----) ( -30.60) >consensus CUCUCUUUUUGGUCCCAUUCUCUCGUCAAAUCGACGAGAGAUCUGCAAUUACAAUCCGUUUGUUGU____GUGCGUGCCAGUGGCGUAUGCAGUGAAUCGUAAGGGGG____G ..((((((.((((.....(((((((((.....))))))))).((((....(((((......)))))....((((((.......))))))))))...)))).))))))...... (-25.36 = -25.52 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:22 2006