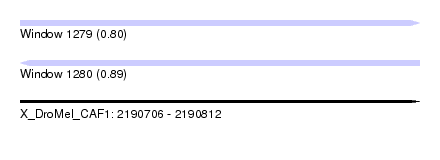
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,190,706 – 2,190,812 |
| Length | 106 |
| Max. P | 0.891123 |

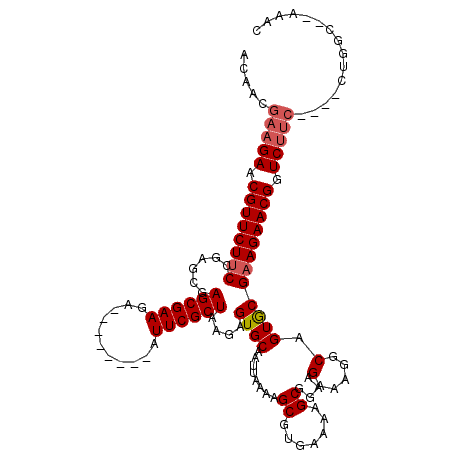
| Location | 2,190,706 – 2,190,812 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.62 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -21.66 |
| Energy contribution | -22.48 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2190706 106 + 22224390 GUUU--GCCAG----GAAGACCGUUCUUCGCACUGCCUUUCUUCCGCUUUUCACGCUUUUAAUUGCACUCUUAGCGAAU--------UCUUCGCUCGCUCGGAAGAACGUUCUUCGUUGU ....--((.(.----(((((.((((((((.(...((..................((........))......(((((..--------...))))).))..))))))))).))))).).)) ( -26.50) >DroSec_CAF1 3721 106 + 1 GUUU--GCCAG----GAAGACCGUUCUUCGCACUGCCUUUCUUCCGCUUUCCACGCUUUUAAUUGCACUGUUAGCGAAU--------UCUUCGCUCGCUCGGCAGAACGUUCUUCGUUGU ....--((.(.----(((((.((((((.......(((.................((........))...((.(((((..--------...))))).))..))))))))).))))).).)) ( -26.11) >DroEre_CAF1 3856 104 + 1 UUUU-UGCCUG-------GACCGUUCUUCGCACUGACUUUCUUCCGCUUUGCACGCUUUUAAGUGCACUCUUAGCGAAU--------UCUUCGCUCGCUCGGAAGAACGUUCUUCGUUGU ....-(((..(-------((....)))..)))..(((.((((((((...(((((........))))).....(((((..--------...)))))....)))))))).)))......... ( -27.30) >DroYak_CAF1 2633 120 + 1 GUUUUUGGAAGGAAGGAAGAGCGUUCUUCGUACCGACUUUCUUCCGCUUUUCACGCUUUUAAUUGCACUCUUAGCGAAUUGGCGAGUUCUUCGCUCGCUCGGAAGAACGUUCUUCGUUGU ............((.((((((((((((((...............((((......((........))......))))....(((((((.....)))))))..)))))))))))))).)).. ( -39.40) >consensus GUUU__GCCAG____GAAGACCGUUCUUCGCACUGACUUUCUUCCGCUUUUCACGCUUUUAAUUGCACUCUUAGCGAAU________UCUUCGCUCGCUCGGAAGAACGUUCUUCGUUGU ...............(((((.((((((((((.......................((........))......((((((...........)))))).))...)))))))).)))))..... (-21.66 = -22.48 + 0.81)

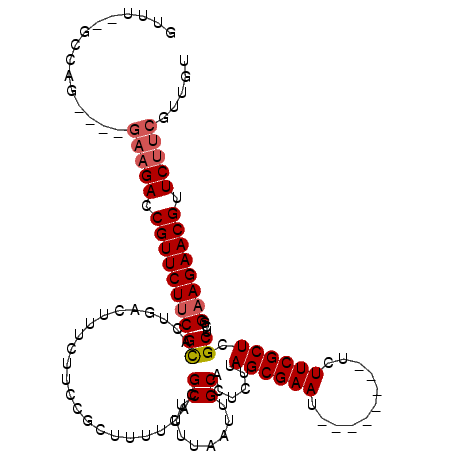
| Location | 2,190,706 – 2,190,812 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.62 |
| Mean single sequence MFE | -29.76 |
| Consensus MFE | -23.81 |
| Energy contribution | -24.62 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.891123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2190706 106 - 22224390 ACAACGAAGAACGUUCUUCCGAGCGAGCGAAGA--------AUUCGCUAAGAGUGCAAUUAAAAGCGUGAAAAGCGGAAGAAAGGCAGUGCGAAGAACGGUCUUC----CUGGC--AAAC .....(((((.((((((((...((((((((...--------..))))).....)))........(((((.....(....).....)).))))))))))).)))))----.....--.... ( -30.00) >DroSec_CAF1 3721 106 - 1 ACAACGAAGAACGUUCUGCCGAGCGAGCGAAGA--------AUUCGCUAACAGUGCAAUUAAAAGCGUGGAAAGCGGAAGAAAGGCAGUGCGAAGAACGGUCUUC----CUGGC--AAAC .....(((((.((((((..((((((((......--------.))))))..((.(((........((.......)).........))).)))).)))))).)))))----.....--.... ( -28.43) >DroEre_CAF1 3856 104 - 1 ACAACGAAGAACGUUCUUCCGAGCGAGCGAAGA--------AUUCGCUAAGAGUGCACUUAAAAGCGUGCAAAGCGGAAGAAAGUCAGUGCGAAGAACGGUC-------CAGGCA-AAAA ........((.((((((((...((..((.....--------.((((((.....(((((........))))).)))))).....))..))..)))))))).))-------......-.... ( -27.40) >DroYak_CAF1 2633 120 - 1 ACAACGAAGAACGUUCUUCCGAGCGAGCGAAGAACUCGCCAAUUCGCUAAGAGUGCAAUUAAAAGCGUGAAAAGCGGAAGAAAGUCGGUACGAAGAACGCUCUUCCUUCCUUCCAAAAAC .....(((((.((((((((((((((((.......)))))...((((((....((((........))))....))))))......))))....))))))).)))))............... ( -33.20) >consensus ACAACGAAGAACGUUCUUCCGAGCGAGCGAAGA________AUUCGCUAAGAGUGCAAUUAAAAGCGUGAAAAGCGGAAGAAAGGCAGUGCGAAGAACGGUCUUC____CUGGC__AAAC .....(((((.((((((((......((((((...........))))))....((((........((.......))....(.....).)))))))))))).)))))............... (-23.81 = -24.62 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:51 2006