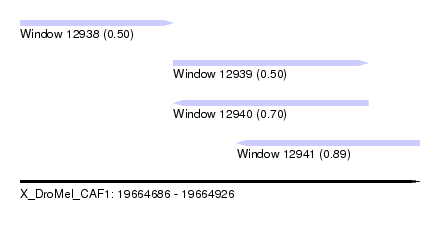
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,664,686 – 19,664,926 |
| Length | 240 |
| Max. P | 0.885259 |

| Location | 19,664,686 – 19,664,778 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 84.28 |
| Mean single sequence MFE | -14.73 |
| Consensus MFE | -9.85 |
| Energy contribution | -10.60 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19664686 92 + 22224390 CUUUAUUUCACUGCCUUACUUAAUUGUUUAUACAUUGAGUUCGAAAUGUGUAAGCAAACAAUACGAAAAC-AAAGAAUACAAAUCAUAUGUAA-- .......................(((((((((((((........))))))))))))).............-......((((.......)))).-- ( -14.00) >DroSec_CAF1 38190 94 + 1 CUUUAAUUCACUGCAUUACUAAAUUGUUUAUACAUUCAGUUCGAAACGUGUAAGCAAACAAAACGGAAAC-AACGGAAACAAAUCAUGUGUAAAA ...............((((....((((((((((.(((.....)))..))))))))))(((....(....)-...(....)......))))))).. ( -15.70) >DroSim_CAF1 29939 85 + 1 CUUUAAUUCACUGCAUUACUUAAUUGUUUAAACAUUCAGUUCGAAACGUGUAAGCAAACAAAACGG----------AAACAAAUCAUGUGUAAAA ........((((((.((((.....(((((.(((.....)))..))))).)))))))........(.----------...).......)))..... ( -10.00) >DroYak_CAF1 52191 93 + 1 AUUUAUUUCACUGCAUUACUUAAUUGUUUAUACAUUGAGUUCGAAAUGUGUAAGCAAACGA-ACGGAAACAAAAGAAUACAAAUAAUGUGUUAA- ............((((((.....(((((((((((((........)))))))))))))....-..(....).............)))))).....- ( -19.20) >consensus CUUUAAUUCACUGCAUUACUUAAUUGUUUAUACAUUCAGUUCGAAACGUGUAAGCAAACAAAACGGAAAC_AAAGAAAACAAAUCAUGUGUAAA_ .......................(((((((((((((........)))))))))))))...................................... ( -9.85 = -10.60 + 0.75)


| Location | 19,664,778 – 19,664,895 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 92.44 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -20.32 |
| Energy contribution | -20.77 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19664778 117 + 22224390 GAUGGCAAACAAAAAUGAUUUAAAAUGCUUUCUGUAGCUAAAGCAUGGAGUAAGUUUCUAGAUUCAAGUUUCUAGAUUUAUCUUCUGCCUUUACUUUAAUAACAUACAUUUCUAUGU ...((((.................(((((((........)))))))((((......((((((........)))))).....))))))))............(((((......))))) ( -22.00) >DroSec_CAF1 38284 109 + 1 UAUGGCAAACAAAAAUGAUUUAAAAUGCUUUCUGUAGCUAAAGCA-GGAGUAAGUUUCUAGAUUCAGGUUUCUAGAUUUAUCUUUUGCCA-----UUAAUAACAUACAUUU--GUGU .((((((((.......((((((...((((((........))))))-....))))))((((((........)))))).......)))))))-----).....(((((....)--)))) ( -23.60) >DroSim_CAF1 30024 110 + 1 GAUGGCAAACAAAAAUGAUUUAAAAUGCUUUCUGUAGCUAAAGCAUGGAGUAAGUUUCUAGAUUCAGGUUUCUAGAUUUAUCUUUUGCCA-----UUAAUAACAUACAUUU--GUGU (((((((((.......((((((..(((((((........)))))))....))))))((((((........)))))).......)))))))-----))....(((((....)--)))) ( -26.10) >consensus GAUGGCAAACAAAAAUGAUUUAAAAUGCUUUCUGUAGCUAAAGCAUGGAGUAAGUUUCUAGAUUCAGGUUUCUAGAUUUAUCUUUUGCCA_____UUAAUAACAUACAUUU__GUGU ...((((((.......((((((..(((((((........)))))))....))))))((((((........)))))).......))))))................((((....)))) (-20.32 = -20.77 + 0.45)



| Location | 19,664,778 – 19,664,895 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 92.44 |
| Mean single sequence MFE | -20.53 |
| Consensus MFE | -18.82 |
| Energy contribution | -18.60 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19664778 117 - 22224390 ACAUAGAAAUGUAUGUUAUUAAAGUAAAGGCAGAAGAUAAAUCUAGAAACUUGAAUCUAGAAACUUACUCCAUGCUUUAGCUACAGAAAGCAUUUUAAAUCAUUUUUGUUUGCCAUC .....(((((((.(((..((((((((..((.(((((.....((((((........))))))..))).)))).))))))))..)))....)))))))..................... ( -20.60) >DroSec_CAF1 38284 109 - 1 ACAC--AAAUGUAUGUUAUUAA-----UGGCAAAAGAUAAAUCUAGAAACCUGAAUCUAGAAACUUACUCC-UGCUUUAGCUACAGAAAGCAUUUUAAAUCAUUUUUGUUUGCCAUA ....--...............(-----(((((((.......((((((........))))))..........-((((((........))))))................)))))))). ( -20.40) >DroSim_CAF1 30024 110 - 1 ACAC--AAAUGUAUGUUAUUAA-----UGGCAAAAGAUAAAUCUAGAAACCUGAAUCUAGAAACUUACUCCAUGCUUUAGCUACAGAAAGCAUUUUAAAUCAUUUUUGUUUGCCAUC ....--...............(-----(((((((.......((((((........))))))..........(((((((........)))))))...............)))))))). ( -20.60) >consensus ACAC__AAAUGUAUGUUAUUAA_____UGGCAAAAGAUAAAUCUAGAAACCUGAAUCUAGAAACUUACUCCAUGCUUUAGCUACAGAAAGCAUUUUAAAUCAUUUUUGUUUGCCAUC ............................((((((.......((((((........))))))...........((((((........))))))................))))))... (-18.82 = -18.60 + -0.22)

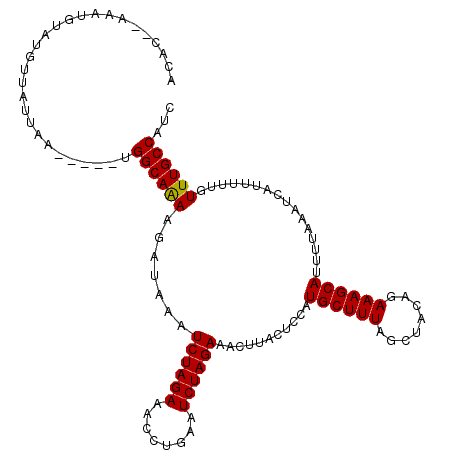

| Location | 19,664,816 – 19,664,926 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 91.95 |
| Mean single sequence MFE | -17.37 |
| Consensus MFE | -15.53 |
| Energy contribution | -15.53 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19664816 110 - 22224390 UAAAUAAAUUUAUAUGAAAUGACACGCAUAUACAUAGAAAUGUAUGUUAUUAAAGUAAAGGCAGAAGAUAAAUCUAGAAACUUGAAUCUAGAAACUUACUCCAUGCUUUA ............(((..(((((((..(((..........)))..)))))))...)))((((((..((.(((.((((((........))))))...)))))...)))))). ( -18.40) >DroSec_CAF1 38322 102 - 1 UAAAUAAAUUUAUAUAAAAUGACACGCAUAUACAC--AAAUGUAUGUUAUUAA-----UGGCAAAAGAUAAAUCUAGAAACCUGAAUCUAGAAACUUACUCC-UGCUUUA .................(((((((..(((......--..)))..)))))))..-----.((((..((.(((.((((((........))))))...)))))..-))))... ( -17.30) >DroSim_CAF1 30062 103 - 1 UAAAUAAAUUUAUAUAAAAUGACACGCAUAUACAC--AAAUGUAUGUUAUUAA-----UGGCAAAAGAUAAAUCUAGAAACCUGAAUCUAGAAACUUACUCCAUGCUUUA .................(((((((..(((......--..)))..))))))).(-----(((...(((.....((((((........))))))..)))...))))...... ( -16.40) >consensus UAAAUAAAUUUAUAUAAAAUGACACGCAUAUACAC__AAAUGUAUGUUAUUAA_____UGGCAAAAGAUAAAUCUAGAAACCUGAAUCUAGAAACUUACUCCAUGCUUUA .................(((((((..(((..........)))..)))))))........((((..((.(((.((((((........))))))...)))))...))))... (-15.53 = -15.53 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:02 2006