
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,658,674 – 19,658,804 |
| Length | 130 |
| Max. P | 0.760948 |

| Location | 19,658,674 – 19,658,764 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 84.88 |
| Mean single sequence MFE | -30.27 |
| Consensus MFE | -27.33 |
| Energy contribution | -27.00 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19658674 90 + 22224390 GGGUU--UUCCGGAAAAGCGGGA----------CCGUUGCAUGCCAACCAGCCAAGCACCAGGACACUUGGCACUGCCCCCCAUUCCAGGACAAUCUGUCCA (((..--(((((......)))))----------.....(((((((((....((........))....)))))).)))..)))......((((.....)))). ( -28.00) >DroEre_CAF1 44809 86 + 1 GGGUU--UCCCGGAAAAGCGGGA----------CCGUUGCAUGCCA-CCAGCUAACCACCCGGACACUUGGCACUGCC-CC--UCCCAGGACAAUCUGUCCA (((..--(((((......)))))----------.....((((((((-..((....((....))...))))))).))).-..--.))).((((.....)))). ( -27.30) >DroYak_CAF1 45046 99 + 1 GGGUUUUUCCCGGAAAAGCGGGAAAGCGGGAAGGCGUUGCAUGCCA-CCAGCUAACCACCCGGACACUUGGCACUGCCCCC--UCCCAGGACAAUCUGUCCA (((.((((((((......)))))))).(((..((((.(((..((..-...))...((....)).......))).)))))))--.))).((((.....)))). ( -35.50) >consensus GGGUU__UCCCGGAAAAGCGGGA__________CCGUUGCAUGCCA_CCAGCUAACCACCCGGACACUUGGCACUGCCCCC__UCCCAGGACAAUCUGUCCA (((..(((((((......))))))).............((((((((.....((........)).....))))).))).......))).((((.....)))). (-27.33 = -27.00 + -0.33)

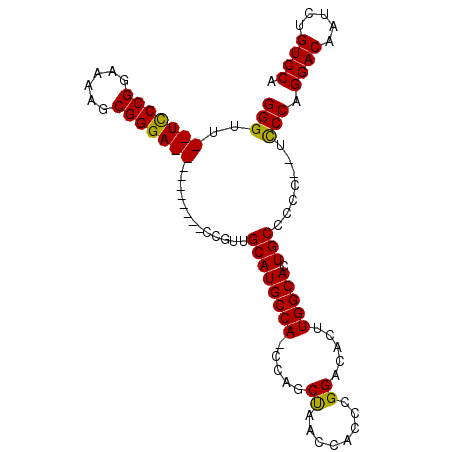

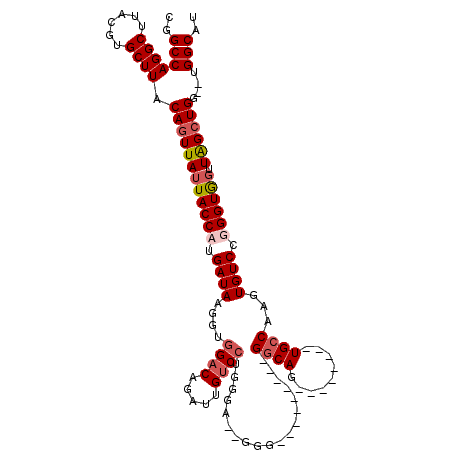
| Location | 19,658,702 – 19,658,804 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -38.48 |
| Consensus MFE | -26.81 |
| Energy contribution | -28.12 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19658702 102 - 22224390 CGGCCAGGCUUACGUGCUUACACUUAUUACCAUGAUAAGGUGGACAGAUUGUCCUGGAAUGGGG---------GGCAG--------UGCCAAGUGUCCUGGUGCUUGGCUGGU-UGGCAU (((((((((....(((....))).....((((.((((...(((....((((((((.......))---------)))))--------).)))..)))).)))))))))))))..-...... ( -38.20) >DroEre_CAF1 44837 98 - 1 CGGCCAGGCUUACGUGCUUACAGUUAUUACCAUGAUAAGGUGGACAGAUUGUCCUGGGA--GG----------GGCAG--------UGCCAAGUGUCCGGGUGGUUAGCUGG--UGGCAU ..(((((((......)))).(((((((((((..((((...(((....((((((((....--))----------)))))--------).)))..))))..))))).)))))).--.))).. ( -36.90) >DroYak_CAF1 45086 99 - 1 CGGCCAGGCUUACGUGCUUACAGUUAUUACCAUGAUAAGGUGGACAGAUUGUCCUGGGA--GGG---------GGCAG--------UGCCAAGUGUCCGGGUGGUUAGCUGG--UGGCAU ..(((((((......)))).(((((((((((..((((...(((....((((((((....--.))---------)))))--------).)))..))))..))))).)))))).--.))).. ( -36.40) >DroAna_CAF1 47811 117 - 1 UGGCCAGGCUUACAUGCUUACAGUUAUUACCAUGAUAAGGUGGACAGAUUGCCAAUAGA--GGGGAGUGGGAGGACAGGCGCGUCCUGGCUAGUGUCCUGGUAG-UGGCUGGUGUGGCAU ..(((((((......)))).(((((((((((.......((..(.....)..)).....(--(((.(.(((.(((((......)))))..))).).)))))))))-))))))....))).. ( -42.40) >consensus CGGCCAGGCUUACGUGCUUACAGUUAUUACCAUGAUAAGGUGGACAGAUUGUCCUGGGA__GGG_________GGCAG________UGCCAAGUGUCCGGGUGGUUAGCUGG__UGGCAU ..(((((((......)))).((((((((((((.((((....((((.....))))...................((((.........))))...)))).)))))).))))))....))).. (-26.81 = -28.12 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:49:58 2006