
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,627,884 – 19,628,001 |
| Length | 117 |
| Max. P | 0.999991 |

| Location | 19,627,884 – 19,628,001 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 62.65 |
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -18.45 |
| Energy contribution | -16.80 |
| Covariance contribution | -1.65 |
| Combinations/Pair | 1.40 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.53 |
| SVM decision value | 5.64 |
| SVM RNA-class probability | 0.999991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
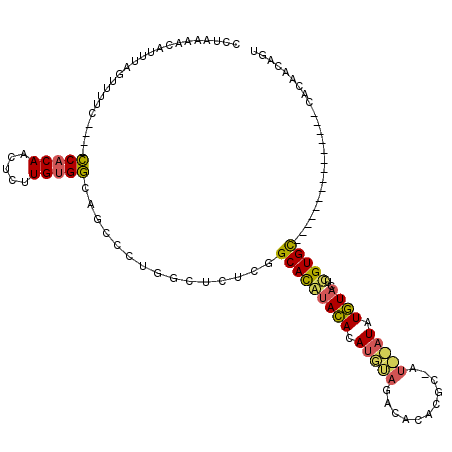
>X_DroMel_CAF1 19627884 117 + 22224390 ACUGU-GUGUACACAUGGGCUUACACACAAAUAUACAUACAUAACAUACAUAUGUAUGUAUGUAUGCAAAUGGCGAGGGUUGUCGCAAUAGUUGUGAUCUGAGUCAUUAAAUAGUUUU-- ..(((-(((((..(....)..))))))))..((((((((((((.(((....)))))))))))))))..((((((..((((..(.((....)).)..))))..))))))..........-- ( -35.20) >DroPse_CAF1 39193 96 + 1 ACUGUUGUG---------------GCACGAGUACAUA----U-GCGUGUGUCUACAUGUGUACGUGCCGAGAGCCAGGGCUGCCACAAGAGUUGUGG----GAAAACUAAAUGUUUUAGG (((.(((((---------------((.((.((((.((----(-((((((....))))))))).))))))..(((....)))))))))).))).....----.(((((.....)))))... ( -35.40) >DroPer_CAF1 39969 100 + 1 ACUGUUGUG---------------GCACGAGUACAUAUGUAU-GCGUGUGUCUACAUGUGUAUGUGCCGAGAGCCAGGGCUGCCACAAGAGUUGUGG----GAAAACUAAAUGCUUUAGG (((.(((((---------------((.((.((((((((....-((((((....))))))))))))))))..(((....)))))))))).))).....----.....((((.....)))). ( -34.30) >consensus ACUGUUGUG_______________GCACGAGUACAUAU__AU_GCGUGUGUCUACAUGUGUAUGUGCCGAGAGCCAGGGCUGCCACAAGAGUUGUGG____GAAAACUAAAUGCUUUAGG (((.(((((.....................((((((((.....((((((....))))))))))))))....(((....)))..))))).)))............................ (-18.45 = -16.80 + -1.65)


| Location | 19,627,884 – 19,628,001 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 62.65 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -8.62 |
| Energy contribution | -8.97 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.37 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19627884 117 - 22224390 --AAAACUAUUUAAUGACUCAGAUCACAACUAUUGCGACAACCCUCGCCAUUUGCAUACAUACAUACAUAUGUAUGUUAUGUAUGUAUAUUUGUGUGUAAGCCCAUGUGUACAC-ACAGU --................................((((......))))....(((((((((((((((....))))).))))))))))...(((((((((.((....)).)))))-)))). ( -31.20) >DroPse_CAF1 39193 96 - 1 CCUAAAACAUUUAGUUUUC----CCACAACUCUUGUGGCAGCCCUGGCUCUCGGCACGUACACAUGUAGACACACGC-A----UAUGUACUCGUGC---------------CACAACAGU ...(((((.....))))).----.....(((.(((((((((((.........)))..(((((.((((........))-)----).)))))...)))---------------))))).))) ( -26.10) >DroPer_CAF1 39969 100 - 1 CCUAAAGCAUUUAGUUUUC----CCACAACUCUUGUGGCAGCCCUGGCUCUCGGCACAUACACAUGUAGACACACGC-AUACAUAUGUACUCGUGC---------------CACAACAGU ...(((((.....))))).----.....(((.(((((((((((.........)))...((((.(((((..(....).-.))))).))))....)))---------------))))).))) ( -24.30) >consensus CCUAAAACAUUUAGUUUUC____CCACAACUCUUGUGGCAGCCCUGGCUCUCGGCACAUACACAUGUAGACACACGC_AU__AUAUGUACUCGUGC_______________CACAACAGU .......................(((((.....)))))...............(((((((((.(((((...........))))).))))..)))))........................ ( -8.62 = -8.97 + 0.35)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:49:51 2006