
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,613,253 – 19,613,392 |
| Length | 139 |
| Max. P | 0.971975 |

| Location | 19,613,253 – 19,613,364 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -45.10 |
| Consensus MFE | -36.33 |
| Energy contribution | -37.08 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19613253 111 + 22224390 CAUACUGGUGGUGAUGGUAGUAGCUAGCGGCGUGGUGCUGGUGCUGCUGCACUGCAGCAUAUU------GCUGU---GCAUGAGCGUGGGCAUGGGCAUGGGCAUAUCCAAAGCCAGCAU .....((.((((..(((.....(((....((.(.(((((..((((..(((((.((((....))------)).))---)))..))))..))))).)))...)))....)))..)))).)). ( -41.90) >DroPse_CAF1 22034 110 + 1 -AUACUGGUGGUGGUGAUAGUAGCUAGCGGCGUGGUGCUGGUGCUGCUGCACUGCAGCAUACU------GCUGG---GCAUGGGCAUGUGCAUGGGCAUGGGCAUAUCCAAAGCCAGCAU -......(((.((((........(((((((..((.(((.(((((....)))))))).))..))------)))))---...(((..((((.(((....))).))))..)))..)))).))) ( -46.80) >DroSec_CAF1 19125 111 + 1 CAUACUGGUGGUGAUGGUAGUAGCUAGCGGCGUGGUGCUGGUGCUGCUGCACUGCAGCAUAUU------GCUGU---GCAUGAGCGUGGGCAUGGGCAUGGGCAUAUCCAAAGCCAGCAU .....((.((((..(((.....(((....((.(.(((((..((((..(((((.((((....))------)).))---)))..))))..))))).)))...)))....)))..)))).)). ( -41.90) >DroAna_CAF1 20452 120 + 1 CAUACUGGUGGUGGUGGUAGUAGCUAGCGGCGUGGUGCUGGUGCUGCUGCACUGCAGCAUACUGGGCAUGCUGGGCGGCAUGAGCGUGUGCAUGGGCAUGGGCAUAUCCAAAGCCAGCAU ....(((((.(..(((.(((((((.(.((((.....)))).)))))))))))..).((((((.(..((((((....))))))..))))))).(((..((....))..)))..)))))... ( -49.80) >consensus CAUACUGGUGGUGAUGGUAGUAGCUAGCGGCGUGGUGCUGGUGCUGCUGCACUGCAGCAUACU______GCUGG___GCAUGAGCGUGGGCAUGGGCAUGGGCAUAUCCAAAGCCAGCAU .....((.((((..(((.....((.....))...(((((.(((((..(((((..((((...........))))....((....))..)))))..))))).)))))..)))..)))).)). (-36.33 = -37.08 + 0.75)

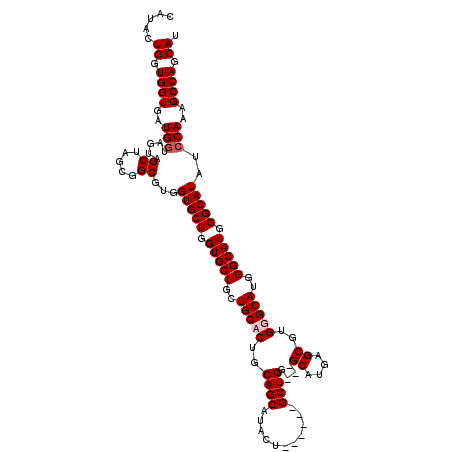

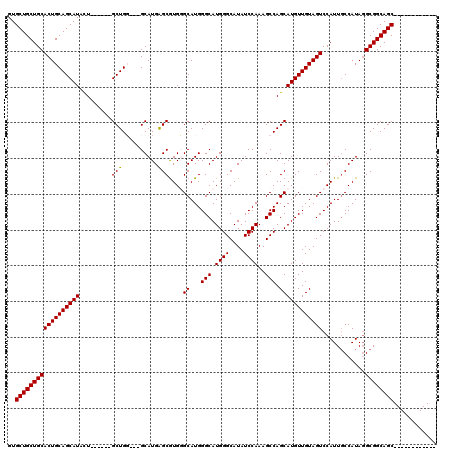
| Location | 19,613,293 – 19,613,392 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.76 |
| Mean single sequence MFE | -45.28 |
| Consensus MFE | -34.71 |
| Energy contribution | -34.52 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19613293 99 + 22224390 GUGCUGCUGCACUGCAGCAUAUU------GCUGU---GCAUGAGCGUGGGCAUGGGCAUGGGCAUAUCCAAAGCCAGCAUGUUGUAGUCCAUUGCCAUAGGCGGCAGC------------ ..((((((((.(((((((.....------)))))---..(((.((((((((...(((((((((.........)))..))))))...))))).))))))))))))))))------------ ( -41.60) >DroPse_CAF1 22073 111 + 1 GUGCUGCUGCACUGCAGCAUACU------GCUGG---GCAUGGGCAUGUGCAUGGGCAUGGGCAUAUCCAAAGCCAGCAUGUUGUAGUCCAUUCCCAUAGGCGGCAGCAGCGACUGCUGC (((((((......))))))).((------(.(((---(.((((((....((((((((.((((....))))..)))..)))))....)))))).)))))))(((((((......))))))) ( -49.70) >DroSec_CAF1 19165 99 + 1 GUGCUGCUGCACUGCAGCAUAUU------GCUGU---GCAUGAGCGUGGGCAUGGGCAUGGGCAUAUCCAAAGCCAGCAUGUUGUAGUCCAUUGCCAUAGGCGGCAGC------------ ..((((((((.(((((((.....------)))))---..(((.((((((((...(((((((((.........)))..))))))...))))).))))))))))))))))------------ ( -41.60) >DroAna_CAF1 20492 108 + 1 GUGCUGCUGCACUGCAGCAUACUGGGCAUGCUGGGCGGCAUGAGCGUGUGCAUGGGCAUGGGCAUAUCCAAAGCCAGCAUGUUGUAGUCCAUUGCCAUAUGCGGCAGC------------ ..((((((((((((((((((.((((.((((((....)))))).((.(((((....))))).))..........)))).))))))))))............))))))))------------ ( -48.20) >consensus GUGCUGCUGCACUGCAGCAUACU______GCUGG___GCAUGAGCGUGGGCAUGGGCAUGGGCAUAUCCAAAGCCAGCAUGUUGUAGUCCAUUGCCAUAGGCGGCAGC____________ ..((((((((((((((((((.........(((..........)))....((...(((.((((....))))..))).))))))))))))............))))))))............ (-34.71 = -34.52 + -0.19)


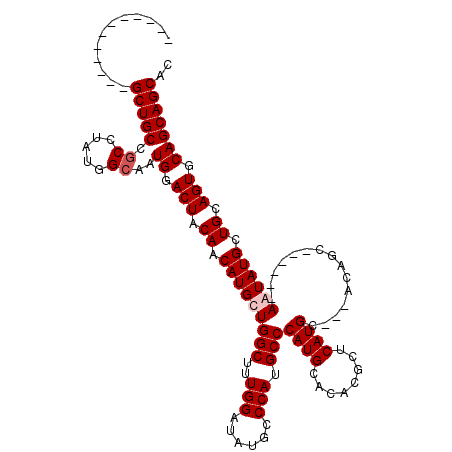
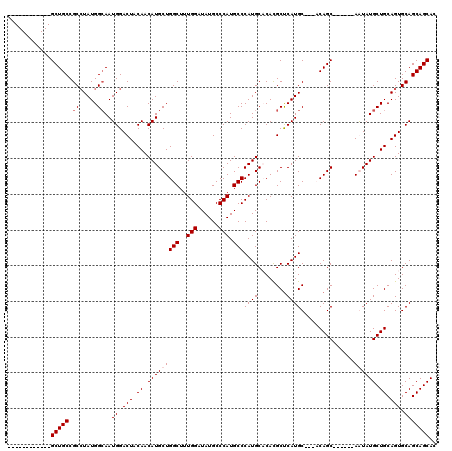
| Location | 19,613,293 – 19,613,392 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.76 |
| Mean single sequence MFE | -38.42 |
| Consensus MFE | -27.23 |
| Energy contribution | -27.97 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19613293 99 - 22224390 ------------GCUGCCGCCUAUGGCAAUGGACUACAACAUGCUGGCUUUGGAUAUGCCCAUGCCCAUGCCCACGCUCAUGC---ACAGC------AAUAUGCUGCAGUGCAGCAGCAC ------------(((((.((....(((((((........)))...(((..(((......))).)))..))))...))...(((---((.((------(......))).)))))))))).. ( -33.50) >DroPse_CAF1 22073 111 - 1 GCAGCAGUCGCUGCUGCCGCCUAUGGGAAUGGACUACAACAUGCUGGCUUUGGAUAUGCCCAUGCCCAUGCACAUGCCCAUGC---CCAGC------AGUAUGCUGCAGUGCAGCAGCAC .........(((((((((.....((((.((((.(.....((((..(((..(((......))).))))))).....).)))).)---)))((------((....)))).).)))))))).. ( -44.70) >DroSec_CAF1 19165 99 - 1 ------------GCUGCCGCCUAUGGCAAUGGACUACAACAUGCUGGCUUUGGAUAUGCCCAUGCCCAUGCCCACGCUCAUGC---ACAGC------AAUAUGCUGCAGUGCAGCAGCAC ------------(((((.((....(((((((........)))...(((..(((......))).)))..))))...))...(((---((.((------(......))).)))))))))).. ( -33.50) >DroAna_CAF1 20492 108 - 1 ------------GCUGCCGCAUAUGGCAAUGGACUACAACAUGCUGGCUUUGGAUAUGCCCAUGCCCAUGCACACGCUCAUGCCGCCCAGCAUGCCCAGUAUGCUGCAGUGCAGCAGCAC ------------(((((.((((..((((.((..(.....((((..(((..(((......))).))))))).....)..))))))...((((((((...))))))))..)))).))))).. ( -42.00) >consensus ____________GCUGCCGCCUAUGGCAAUGGACUACAACAUGCUGGCUUUGGAUAUGCCCAUGCCCAUGCACACGCUCAUGC___ACAGC______AAUAUGCUGCAGUGCAGCAGCAC ............(((((.((.....))..((.(((.((.(((((((((..(((......))).)))((((........))))...............)))))).)).))).))))))).. (-27.23 = -27.97 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:49:48 2006