
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,611,958 – 19,612,278 |
| Length | 320 |
| Max. P | 0.966454 |

| Location | 19,611,958 – 19,612,078 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.83 |
| Mean single sequence MFE | -48.74 |
| Consensus MFE | -39.38 |
| Energy contribution | -39.62 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19611958 120 - 22224390 GCCGCAUAAGUGCGAGCUGUGCCCCAAGCAGUUCCAUCACAAGACGGAUCUGCGCCGACACGUGGAGGCCAUCCAUACGGGGCUGAAGCAGCACAUGUGCGACAUCUGUGAGAAGGGCUU ((((((((.((((..(((..(((((..((((.(((.((....)).))).))))(((..(....)..))).........)))))...))).)))).))))).....((......))))).. ( -52.10) >DroPse_CAF1 20546 120 - 1 GCCGCACAAGUGCGAGCUCUGUCCGAAGCAGUUCCAUCACAAGACCGAUCUGCGGCGCCACGUGGAGGCCAUCCACACGGGGCUCAAGCAGCAUAUGUGCGACAUUUGCGAGAAGGGCUU ((((((((.(((((((((((((.((..((((.((..((....))..)).))))..))....(((((.....)))))))))))))).....)))).)))))........(.....)))).. ( -45.00) >DroSec_CAF1 17711 120 - 1 GCCGCACAAGUGCGAGCUGUGCCCCAAGCAGUUCCACCACAAGACGGAUCUGCGCCGACACGUGGAGGCCAUCCACACGGGGCUGAAGCAGCACAUGUGCGACAUCUGCGAGAAGGGCUU ((((((((.((((..(((..(((((..((((.(((..........))).))))........(((((.....)))))..)))))...))).)))).))))).....((......))))).. ( -53.60) >DroAna_CAF1 18983 120 - 1 GCCGCACAAGUGCGACCAGUGCCCCAAGCAGUUCCACCACAAAACCGACCUGCGCCGGCACGUCGAGGCCAUCCACACGGGCCUCAAGCAGCACAUGUGCGACAUCUGCGAGAAGGGCUU ((((((((.(((((((..(((((....((((.((............)).))))...))))))))((((((.........)))))).....)))).))))).....((......))))).. ( -48.00) >DroPer_CAF1 21453 120 - 1 GCCGCACAAGUGCGAGCUCUGUCCGAAGCAGUUCCAUCACAAGACCGAUCUGCGGCGCCACGUGGAGGCCAUCCACACGGGGCUCAAGCAGCAUAUGUGCGACAUUUGCGAGAAGGGCUU ((((((((.(((((((((((((.((..((((.((..((....))..)).))))..))....(((((.....)))))))))))))).....)))).)))))........(.....)))).. ( -45.00) >consensus GCCGCACAAGUGCGAGCUGUGCCCCAAGCAGUUCCAUCACAAGACCGAUCUGCGCCGACACGUGGAGGCCAUCCACACGGGGCUCAAGCAGCACAUGUGCGACAUCUGCGAGAAGGGCUU ((((((((.((((..(((..(((((..((((.((............)).))))........(((((.....)))))..)))))...))).)))).))))).....((......))))).. (-39.38 = -39.62 + 0.24)


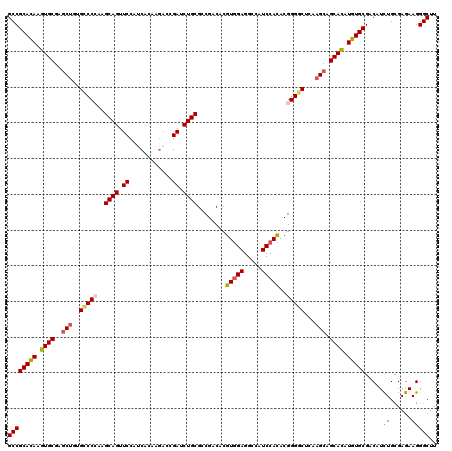
| Location | 19,612,038 – 19,612,158 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.83 |
| Mean single sequence MFE | -47.72 |
| Consensus MFE | -37.70 |
| Energy contribution | -38.10 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.927869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19612038 120 - 22224390 CGCUGCCAGAAGUGCGGCAAGCGCUUUCAGAGCGAGGAUGUCUACACCACGCACCUGGGUCGCCACCGCACCCAGGACAAGCCGCAUAAGUGCGAGCUGUGCCCCAAGCAGUUCCAUCAC .(((((..(..(..((((..((((((...(.(((.((.((....)))).))).(((((((.((....)))))))))......)....))))))..))))..)..)..)))))........ ( -47.10) >DroPse_CAF1 20626 120 - 1 CGCUGCCAGAAGUGCGGCAAGCGCUUCCAGAGCGAGGACGUUUACACGACACACCUGGGCCGGCACCGUACCCAGGACAAGCCGCACAAGUGCGAGCUCUGUCCGAAGCAGUUCCAUCAC .(((((..(((((((.....)))))))((((((..((.(((....))).....((((((.((....))..)))))).....))(((....)))..))))))......)))))........ ( -45.90) >DroSec_CAF1 17791 120 - 1 CGCUGCCAGAAGUGCGGCAAGCGCUUUCAGAGCGAGGAUGUCUACACCACGCACCUGGGUCGCCACCGCACCCAGGAUAAGCCGCACAAGUGCGAGCUGUGCCCCAAGCAGUUCCACCAC ...........(((((((.............(((.((.((....)))).))).(((((((.((....)))))))))....)))))))..(((.(((((((.......))))))))))... ( -50.20) >DroAna_CAF1 19063 120 - 1 CGGUGCCAGAAGUGCGGCAAGCGCUUCCAGAGCGAAGACGUCUACACGACGCACCUGGGGCGCCACCGCACCCAGGACAAGCCGCACAAGUGCGACCAGUGCCCCAAGCAGUUCCACCAC .((((......(((((((...(((((...)))))..(.((((.....))))).(((((((((....))).))))))....)))))))..(..(.....)..)............)))).. ( -48.50) >DroPer_CAF1 21533 120 - 1 CGCUGCCAGAAGUGCGGCAAGCGCUUCCAGAGCGAGGAUGUUUACACGACACACCUGGGCCGGCACCGUACCCAGGACAAGCCGCACAAGUGCGAGCUCUGUCCGAAGCAGUUCCAUCAC ((((.(..(((((((.....)))))))..))))).((((((((....((((..((((((.((....))..))))))...(((((((....)))).))).))))..)))))..)))..... ( -46.90) >consensus CGCUGCCAGAAGUGCGGCAAGCGCUUCCAGAGCGAGGAUGUCUACACGACGCACCUGGGCCGCCACCGCACCCAGGACAAGCCGCACAAGUGCGAGCUGUGCCCCAAGCAGUUCCAUCAC ...........(((((((...(((((...)))))...................((((((...........))))))....)))))))..(((.(((((((.......))))))))))... (-37.70 = -38.10 + 0.40)

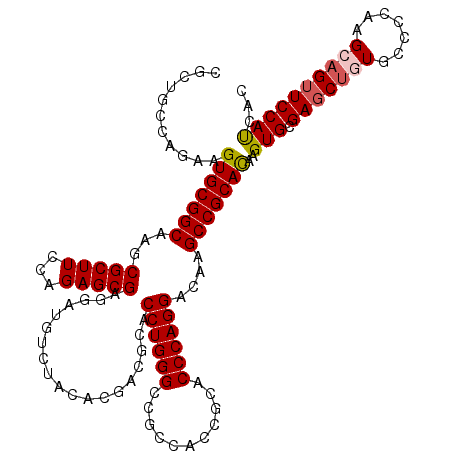
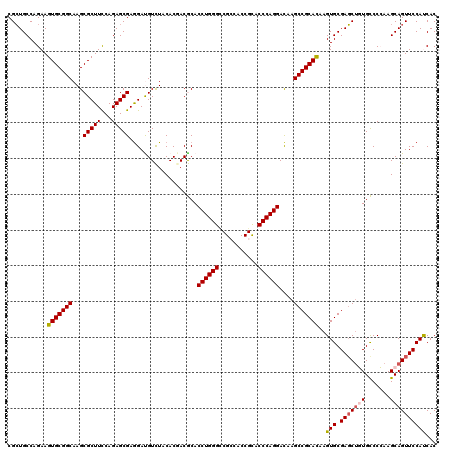
| Location | 19,612,078 – 19,612,198 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -43.08 |
| Consensus MFE | -36.00 |
| Energy contribution | -35.60 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19612078 120 - 22224390 GCAGCACAACAAGAGCUUCCACUCCGGUGAGUACCCAUUUCGCUGCCAGAAGUGCGGCAAGCGCUUUCAGAGCGAGGAUGUCUACACCACGCACCUGGGUCGCCACCGCACCCAGGACAA ...((.......(((......))).((((((.((...(((((((.(..(((((((.....)))))))..))))))))..)))).))))..)).(((((((.((....))))))))).... ( -43.60) >DroPse_CAF1 20666 120 - 1 GCAGCACAACAAGAGCUUCCACUCCGGUGAGUACCCGUUCCGCUGCCAGAAGUGCGGCAAGCGCUUCCAGAGCGAGGACGUUUACACGACACACCUGGGCCGGCACCGUACCCAGGACAA ....(((.....(((......)))..))).(((..(((((((((.(..(((((((.....)))))))..))))).)))))..)))........((((((.((....))..)))))).... ( -42.50) >DroSec_CAF1 17831 120 - 1 GCAGCACAACAAGAGCUUCCACUCCGGUGAGUACCCGUUUCGCUGCCAGAAGUGCGGCAAGCGCUUUCAGAGCGAGGAUGUCUACACCACGCACCUGGGUCGCCACCGCACCCAGGAUAA ...((.......(((......))).((((((.((...(((((((.(..(((((((.....)))))))..))))))))..)))).))))..)).(((((((.((....))))))))).... ( -44.70) >DroAna_CAF1 19103 120 - 1 GCAGCACAACAAGAGCUUCCACUCCGGUGAGUACCCGUUCCGGUGCCAGAAGUGCGGCAAGCGCUUCCAGAGCGAAGACGUCUACACGACGCACCUGGGGCGCCACCGCACCCAGGACAA ...((.........(((((..(((.(.((.(((((......)))))))(((((((.....)))))))).))).)))).)(((.....))))).(((((((((....))).)))))).... ( -43.90) >DroPer_CAF1 21573 120 - 1 GCAGCACAACAAGAGCUUCCACUCCGGUGAGUACCCGUUCCGCUGCCAGAAGUGCGGCAAGCGCUUCCAGAGCGAGGAUGUUUACACGACACACCUGGGCCGGCACCGUACCCAGGACAA ....(((.....(((......)))..))).(((..(((((((((.(..(((((((.....)))))))..))))).)))))..)))........((((((.((....))..)))))).... ( -40.70) >consensus GCAGCACAACAAGAGCUUCCACUCCGGUGAGUACCCGUUCCGCUGCCAGAAGUGCGGCAAGCGCUUCCAGAGCGAGGAUGUCUACACGACGCACCUGGGCCGCCACCGCACCCAGGACAA ....(((.....(((......)))..))).(((..(((((((((.(..(((((((.....)))))))..))))).)))))..)))........((((((...........)))))).... (-36.00 = -35.60 + -0.40)

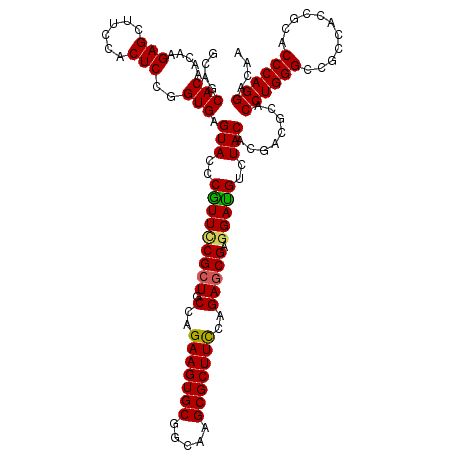

| Location | 19,612,158 – 19,612,278 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -39.72 |
| Consensus MFE | -34.30 |
| Energy contribution | -34.50 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19612158 120 - 22224390 CCGCACUCGACCACCCUGCCAUCGGGACGGAUCAAGUGUCUGGAGUGCGACAAGGAGUUCACCAAGAACUGCUACCUCACGCAGCACAACAAGAGCUUCCACUCCGGUGAGUACCCAUUU .(((((((.....(((.......))).(((((.....))))))))))))....((..((((((.....((((........))))........(((......))).))))))..))..... ( -37.40) >DroPse_CAF1 20746 120 - 1 CCGCACUCGACAACGCUGCCCUCGGGGCGGAUCAAGUGUCUGGAGUGCGACAAGGAGUUCACCAAGAACUGCUACCUCACGCAGCACAACAAGAGCUUCCACUCCGGUGAGUACCCGUUC .(((((((((((...((((((...))))))......))))..)))))))....((..((((((.....((((........))))........(((......))).))))))..))..... ( -42.40) >DroSec_CAF1 17911 120 - 1 CCGCACUCGACCACCCUGCCCUCGGGACGGAUCAAGUGUCUGGAGUGCGACAAGGAGUUCACCAAGAACUGCUACCUCACGCAGCACAACAAGAGCUUCCACUCCGGUGAGUACCCGUUU .(((((((.....(((.......))).(((((.....))))))))))))....((..((((((.....((((........))))........(((......))).))))))..))..... ( -37.40) >DroAna_CAF1 19183 120 - 1 CCGCACUCGACCACGCUGCCGUCGGGCAGGAUCAAGUGUCUGGAGUGCGACAAGGAGUUCACCAAGAACUGCUACCUCACGCAGCACAACAAGAGCUUCCACUCCGGUGAGUACCCGUUC .(((((((.(.(((.(((((....)))))......)))..).)))))))....((..((((((.....((((........))))........(((......))).))))))..))..... ( -39.00) >DroPer_CAF1 21653 120 - 1 CCGCACUCGACAACGCUGCCCUCCGGGCGGAUCAAGUGUCUGGAGUGCGACAAGGAGUUCACCAAGAACUGCUACCUCACGCAGCACAACAAGAGCUUCCACUCCGGUGAGUACCCGUUC .(((((((((((...((((((...))))))......))))..)))))))....((..((((((.....((((........))))........(((......))).))))))..))..... ( -42.40) >consensus CCGCACUCGACCACGCUGCCCUCGGGACGGAUCAAGUGUCUGGAGUGCGACAAGGAGUUCACCAAGAACUGCUACCUCACGCAGCACAACAAGAGCUUCCACUCCGGUGAGUACCCGUUC .((((((((((.((.(((((....)).))).....)))))..)))))))....((..((((((.....((((........))))........(((......))).))))))..))..... (-34.30 = -34.50 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:49:44 2006