
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,608,105 – 19,608,244 |
| Length | 139 |
| Max. P | 0.882785 |

| Location | 19,608,105 – 19,608,214 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.10 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -18.99 |
| Energy contribution | -18.83 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
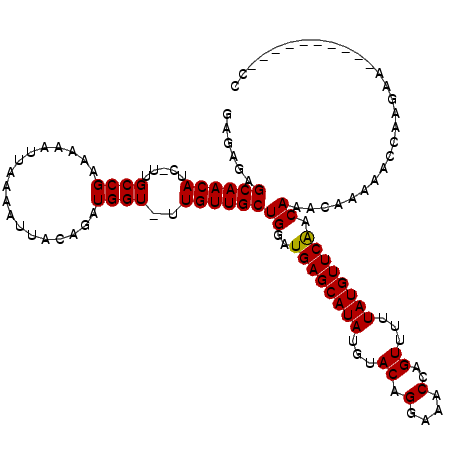
>X_DroMel_CAF1 19608105 109 + 22224390 CUGAGCGCAACAUC-UUGCCGAAAAAUUAAAAUUACAGAUGGU-UUGUUGCUGGAUGAGCAUAUGUACAGGAAACCAGUUUUUAUGUUCAACAAACAAAAACCAAGAA--------A-CC ....(.((((....-)))))...................((((-((.(((...(.((((((((...((.(....)..))...)))))))).)...)))))))))....--------.-.. ( -20.50) >DroPse_CAF1 16202 107 + 1 GAGAGAGCAACAUCGUUGCCGAAAAAUUAAAAUUACAGAUGGUUUUGUUGCUGGAUGAGCAUAUGUACAGGAAACCAGUUUUUAUGUUCAACAAACAAA-ACCAAGAA------------ ....(.(((((...))))))...................((((((((((....(.((((((((...((.(....)..))...)))))))).).))))))-))))....------------ ( -27.50) >DroSec_CAF1 13919 117 + 1 CUGAGCGCAACAUC-UUGCCGAAAAAUUAAAAUUACAGAUGGU-UUGUUGCUGGAUGAGCAUAUGUACAGGAAACCAGUUUUUAUGUUCAACAAACAAAAACCAAGAAACCAAGAAA-CC ....(.((((....-)))))...................((((-((.(((...(.((((((((...((.(....)..))...)))))))).)...))))))))).............-.. ( -20.50) >DroAna_CAF1 16397 117 + 1 GGGAGGGCAACAUC-UUGCCGAAAAAUUAAAAUUACAGAUGGU-UUGUUGCUGGAUGAGCAUAUGUACAGGAAACCAGUUUUUAUGUUCGACAAACACCA-GAAAGAACACAAGAACUCC .(((((((((....-)))))...................((((-((....((((..(((((((...((.(....)..))...)))))))(.....).)))-)...)))).))....)))) ( -28.00) >DroPer_CAF1 17018 107 + 1 GAGAGAGCAACAUCGUUGCCGAAAAAUUAAAAUUACAGAUGGUUUUGUUGCUGGAUGAGCAUAUGUACAGGAAACCAGUUUUUAUGUUCAACAAACAAA-ACCAAGAA------------ ....(.(((((...))))))...................((((((((((....(.((((((((...((.(....)..))...)))))))).).))))))-))))....------------ ( -27.50) >consensus GAGAGAGCAACAUC_UUGCCGAAAAAUUAAAAUUACAGAUGGU_UUGUUGCUGGAUGAGCAUAUGUACAGGAAACCAGUUUUUAUGUUCAACAAACAAAAACCAAGAA__________CC ......((((((.....((((..................))))..))))))((..((((((((...((.(....)..))...)))))))).))........................... (-18.99 = -18.83 + -0.16)

| Location | 19,608,144 – 19,608,244 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.77 |
| Mean single sequence MFE | -25.19 |
| Consensus MFE | -15.14 |
| Energy contribution | -15.90 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19608144 100 + 22224390 GGU-UUGUUGCUGGAUGAGCAUAUGUACAGGAAACCAGUUUUUAUGUUCAACAAACAAAAACCAAGAA--------A-CCAAGA--AACGCACGGGCACCGAAA--AUCGAG-----UA- (((-((.(((...(.((((((((...((.(....)..))...)))))))).)...)))))))).....--------.-......--...((....))((((...--..)).)-----).- ( -19.40) >DroPse_CAF1 16242 93 + 1 GGUUUUGUUGCUGGAUGAGCAUAUGUACAGGAAACCAGUUUUUAUGUUCAACAAACAAA-ACCAAGAA---------------A--AACGCACGA---ACGAGA--AUCGAGUG---UA- (((((((((....(.((((((((...((.(....)..))...)))))))).).))))))-))).....---------------.--.((((.(((---......--.))).)))---).- ( -27.60) >DroSec_CAF1 13958 108 + 1 GGU-UUGUUGCUGGAUGAGCAUAUGUACAGGAAACCAGUUUUUAUGUUCAACAAACAAAAACCAAGAAACCAAGAAA-CCAAGA--AACGCACGGGCACCGAAA--ACCGAG-----UA- (((-((.(((...(.((((((((...((.(....)..))...)))))))).)...))))))))..............-......--...((.(((.........--.))).)-----).- ( -21.30) >DroAna_CAF1 16436 118 + 1 GGU-UUGUUGCUGGAUGAGCAUAUGUACAGGAAACCAGUUUUUAUGUUCGACAAACACCA-GAAAGAACACAAGAACUCCAAGAAAAACGCACGGCAGCCGAAAGAAUCGCGCGCCGUAG (((-((.(((.((.(((....))).))))).)))))(((((((.(((((...........-....))))).))))))).............(((((.(((((.....))).))))))).. ( -30.06) >DroPer_CAF1 17058 93 + 1 GGUUUUGUUGCUGGAUGAGCAUAUGUACAGGAAACCAGUUUUUAUGUUCAACAAACAAA-ACCAAGAA---------------A--AACGCACGA---ACGAGA--AUCGAGUG---UA- (((((((((....(.((((((((...((.(....)..))...)))))))).).))))))-))).....---------------.--.((((.(((---......--.))).)))---).- ( -27.60) >consensus GGU_UUGUUGCUGGAUGAGCAUAUGUACAGGAAACCAGUUUUUAUGUUCAACAAACAAAAACCAAGAA__________CCAAGA__AACGCACGG___CCGAAA__AUCGAG_G___UA_ (((..((((....(.((((((((...((.(....)..))...)))))))).).))))...))).........................(((.(((............))).)))...... (-15.14 = -15.90 + 0.76)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:49:40 2006