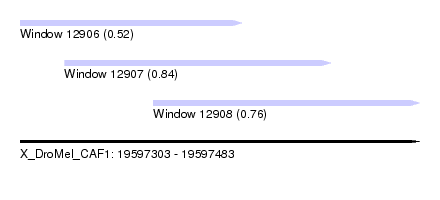
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,597,303 – 19,597,483 |
| Length | 180 |
| Max. P | 0.844466 |

| Location | 19,597,303 – 19,597,403 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.80 |
| Mean single sequence MFE | -32.93 |
| Consensus MFE | -27.61 |
| Energy contribution | -27.05 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
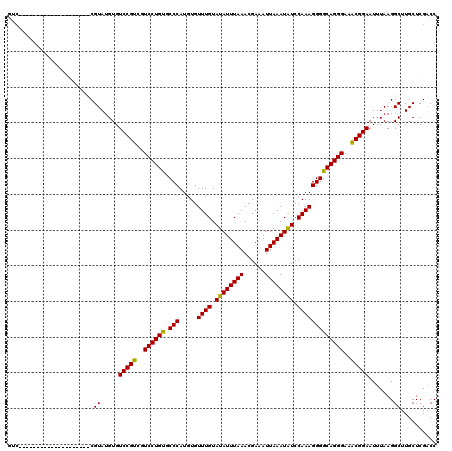
>X_DroMel_CAF1 19597303 100 + 22224390 GUC--------------------CGUAUGUGUCCGUCGUCCUGUACCCAUGUGUUUGUAUAUUUAAACGAAAUUAAAUGUCCAAAGGGGCAGGGAAACGGAAUUUAAGGCUUGCUCGACC (((--------------------.(((.((.(((((..((((((.(((.....((((.((((((((......)))))))).)))))))))))))..))))).......)).)))..))). ( -28.50) >DroSec_CAF1 3653 120 + 1 GUCCGUGUGUGUCCGUGUGCGUCCGUAUGUGUCCGUCGUCCUGUGCCCAUGUGUUUGUAUAUUUAAACGAAAUUAAAUAUCCAAAGGGGCAGGGAAACGGAAUUUAAGGCUUGCUCGCCC ...((.(((.(((((..(((....)))..))(((((..((((((.(((.....((((.((((((((......)))))))).)))))))))))))..)))))......))).))).))... ( -35.40) >DroSim_CAF1 3616 100 + 1 GUC--------------------CGUAUGUGUCCGUCGUCCUGUGCCCAUGUGUUUGUAUAUUUAAACGAAAUUAAAUAUCCAAAGGGGCAGGGAUACGGAAUUUAAGGCUUGCUCGACC (((--------------------.(((.((.(((((.(((((.(((((.....((((.((((((((......)))))))).)))).))))))))))))))).......)).)))..))). ( -30.00) >DroEre_CAF1 3891 109 + 1 -----------UCCGUGUGUGUCCGUAUGUGUCCGUCGUCCUGCUCCCAUGUGUUUGUAUAUUUAAACGAAAUUAAAUAUCCAAAGGGGCAGGGAGGCGGAAUUUAAGGCUUGCUCGACC -----------..((.(((.(.((.......((((((..((((((((......((((.((((((((......)))))))).))))))))))))..))))))......))).))).))... ( -37.82) >consensus GUC____________________CGUAUGUGUCCGUCGUCCUGUGCCCAUGUGUUUGUAUAUUUAAACGAAAUUAAAUAUCCAAAGGGGCAGGGAAACGGAAUUUAAGGCUUGCUCGACC ........................((.....(((((..((((((.(((.....((((.((((((((......)))))))).)))))))))))))..))))).......)).......... (-27.61 = -27.05 + -0.56)


| Location | 19,597,323 – 19,597,443 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -34.08 |
| Consensus MFE | -28.34 |
| Energy contribution | -28.27 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
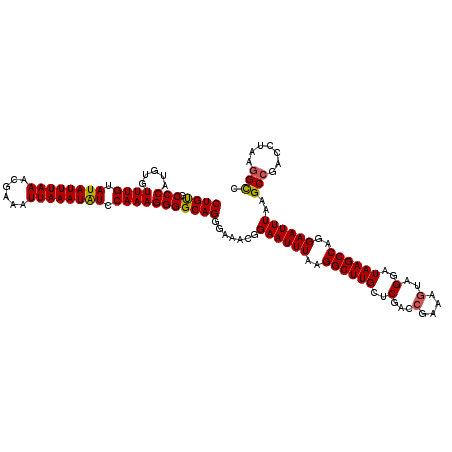
>X_DroMel_CAF1 19597323 120 + 22224390 CUGUACCCAUGUGUUUGUAUAUUUAAACGAAAUUAAAUGUCCAAAGGGGCAGGGAAACGGAAUUUAAGGCUUGCUCGACCGAAUAUAGGAUAAGCCAGGAAUUUAAGCCAACCUAAGGCC ((((.(((.....((((.((((((((......)))))))).)))))))))))(....).((((((..((((((..(...........)..))))))..))))))..(((.......))). ( -30.10) >DroSec_CAF1 3693 120 + 1 CUGUGCCCAUGUGUUUGUAUAUUUAAACGAAAUUAAAUAUCCAAAGGGGCAGGGAAACGGAAUUUAAGGCUUGCUCGCCCAAAAGUAGGAUAAGCCAGGAAUUUAAGCCGACCUAAGGCC ((.(((((.....((((.((((((((......)))))))).)))).))))).)).....((((((..((((((((..(......)..)).))))))..))))))..(((.......))). ( -35.00) >DroSim_CAF1 3636 120 + 1 CUGUGCCCAUGUGUUUGUAUAUUUAAACGAAAUUAAAUAUCCAAAGGGGCAGGGAUACGGAAUUUAAGGCUUGCUCGACCGAAAGUAGGAUAAGCCAGGAAUUUAAGCCGACCCAAGGCC ((.(((((.....((((.((((((((......)))))))).)))).))))).)).....((((((..((((((..(...(....)..)..))))))..))))))..(((.......))). ( -36.40) >DroEre_CAF1 3920 120 + 1 CUGCUCCCAUGUGUUUGUAUAUUUAAACGAAAUUAAAUAUCCAAAGGGGCAGGGAGGCGGAAUUUAAGGCUUGCUCGACCGAAAGUAGGAUAAGCCAGGAAUUUAAGCAGACCUAAGGUU (((((((......((((.((((((((......)))))))).)))))))))))((..((.((((((..((((((..(...(....)..)..))))))..))))))..))...))....... ( -34.80) >consensus CUGUGCCCAUGUGUUUGUAUAUUUAAACGAAAUUAAAUAUCCAAAGGGGCAGGGAAACGGAAUUUAAGGCUUGCUCGACCGAAAGUAGGAUAAGCCAGGAAUUUAAGCCGACCUAAGGCC ((((.(((.....((((.((((((((......)))))))).))))))))))).......((((((..((((((..(...(....)..)..))))))..))))))..(((.......))). (-28.34 = -28.27 + -0.06)


| Location | 19,597,363 – 19,597,483 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.06 |
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -24.19 |
| Energy contribution | -23.88 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
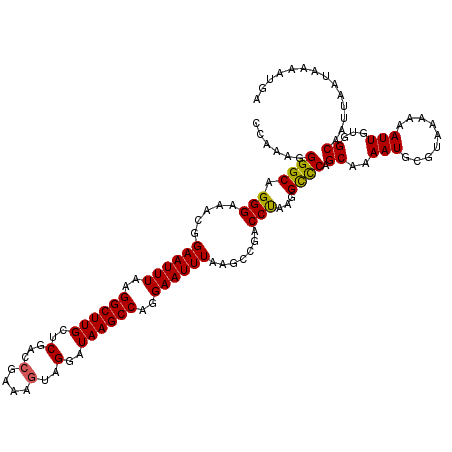
>X_DroMel_CAF1 19597363 120 + 22224390 CCAAAGGGGCAGGGAAACGGAAUUUAAGGCUUGCUCGACCGAAUAUAGGAUAAGCCAGGAAUUUAAGCCAACCUAAGGCCCAGCAAAAUGGGUAAAAAAUUGUGGCAAUUAAUAAAAUGU (((.((((((..(....).((((((..((((((..(...........)..))))))..))))))..)))..)))...(((((......))))).........)))............... ( -30.10) >DroSec_CAF1 3733 120 + 1 CCAAAGGGGCAGGGAAACGGAAUUUAAGGCUUGCUCGCCCAAAAGUAGGAUAAGCCAGGAAUUUAAGCCGACCUAAGGCCCAGCAAAAUGCGUAAAAAAUUGUGGCAAUUAAUAAAAUGA (((...((((.(((...((((((((..((((((((..(......)..)).))))))..)))))....))).)))...)))).((.....))...........)))............... ( -28.70) >DroSim_CAF1 3676 120 + 1 CCAAAGGGGCAGGGAUACGGAAUUUAAGGCUUGCUCGACCGAAAGUAGGAUAAGCCAGGAAUUUAAGCCGACCCAAGGCCCAGCAAAAUGCGUAAAAAAUUGUGGCAAUUAAUAAAAUGA (((...((((.(((...((((((((..((((((..(...(....)..)..))))))..)))))....))).)))...)))).((.....))...........)))............... ( -32.00) >DroEre_CAF1 3960 120 + 1 CCAAAGGGGCAGGGAGGCGGAAUUUAAGGCUUGCUCGACCGAAAGUAGGAUAAGCCAGGAAUUUAAGCAGACCUAAGGUUCAGCAAAAUGGGUACAAAAUUGUUGCAAUUAAUAAAAUGG (((.......(((...((.((((((..((((((..(...(....)..)..))))))..))))))..))...)))..((((((((((..((....))...)))))).)))).......))) ( -28.20) >consensus CCAAAGGGGCAGGGAAACGGAAUUUAAGGCUUGCUCGACCGAAAGUAGGAUAAGCCAGGAAUUUAAGCCGACCUAAGGCCCAGCAAAAUGCGUAAAAAAUUGUGGCAAUUAAUAAAAUGA ......((((.(((.....((((((..((((((..(...(....)..)..))))))..)))))).......)))...)))).((..(((.........)))...)).............. (-24.19 = -23.88 + -0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:49:33 2006