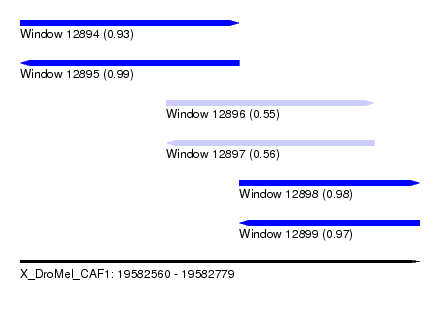
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,582,560 – 19,582,779 |
| Length | 219 |
| Max. P | 0.991342 |

| Location | 19,582,560 – 19,582,680 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.67 |
| Mean single sequence MFE | -44.70 |
| Consensus MFE | -38.28 |
| Energy contribution | -38.36 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.927974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19582560 120 + 22224390 UUGUCGCGGUUGCGCGCGCAACUCGCAACGGUCUGGUAUCGAUCACAGCUGCGAGCAGGCUGCCAGAUCGCUUCCCGUUCGGGAACGAUUUCGCAUCGAUUUUCGAUUGCUGGCCGGUAU .....((((((((....))))).)))..(((.(..(((((((.....(.(((((((.....))..(((((.(((((....))))))))))))))).).....)))).)))..)))).... ( -47.70) >DroSec_CAF1 2481 120 + 1 UUGUCGCGGUUGCGCGCGCAAUUCGCAACGGUCUGUUAUCGAUCACAGCCGCGAGCAGGCUGCCAGAUCGCUUCCCGUAUGGGAACGAUUUCGCAUCAAUUUUCGAUUGCUGACCGGUAU .....((((((((....)))).))))..(((((.((.(((((....((((.......))))((.((((((.(((((....))))))))))).))........))))).)).))))).... ( -47.40) >DroSim_CAF1 2487 120 + 1 UUGUCGCGGUUGCGCGCGCAAUUCGCAACGGUCUGUUAUCGAUCACAGCCGCGAGCAGGCUGCCAGAUCGCUUCCCGUAUGGGAACGAUUUCGCAUCAAUUUUCGAUUGCUGGCCGGUAU .....((((((((....)))).))))..(((((.((.(((((....((((.......))))((.((((((.(((((....))))))))))).))........))))).)).))))).... ( -46.80) >DroEre_CAF1 2438 120 + 1 UUGUGGCGGUUGCGCGCGCAACUCGCAACGGUCUGUUAUCGAUCACAGCUGCGAACAUGCUGCCAGAUCACCUCUUCUAUGAAAAUGAUACUGUAUCGAUUUCCGAUUGCUGGCCGGUAU .....((((((((....))))).)))..(((((.((.(((((((.((((((....)).))))...))))...........((((.(((((...))))).)))).))).)).))))).... ( -36.80) >DroYak_CAF1 2326 120 + 1 UUGUUGCGGUUGCGCGCGCAACUCGCAACGGUCUGUUAUCGAUCACAGCUGCGAGCAGGCUGCCAGACUGCUCCCCUUAUGGGAACAAUUCUGUUUCGAUUUCCGAUUGCUGGCCAGUAU ..(((((((((((....))))).))))))((((.((.(((((((((((....((((((.........))))))(((....))).......))))...)))...)))).)).))))..... ( -44.80) >consensus UUGUCGCGGUUGCGCGCGCAACUCGCAACGGUCUGUUAUCGAUCACAGCUGCGAGCAGGCUGCCAGAUCGCUUCCCGUAUGGGAACGAUUUCGCAUCGAUUUUCGAUUGCUGGCCGGUAU .....((((((((....))))).)))..(((((.((.((((.....((((.......))))((.((((((.(((((....))))))))))).)).........)))).)).))))).... (-38.28 = -38.36 + 0.08)

| Location | 19,582,560 – 19,582,680 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.67 |
| Mean single sequence MFE | -44.46 |
| Consensus MFE | -36.90 |
| Energy contribution | -35.50 |
| Covariance contribution | -1.40 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19582560 120 - 22224390 AUACCGGCCAGCAAUCGAAAAUCGAUGCGAAAUCGUUCCCGAACGGGAAGCGAUCUGGCAGCCUGCUCGCAGCUGUGAUCGAUACCAGACCGUUGCGAGUUGCGCGCGCAACCGCGACAA ....((((((((.((((.....))))((((.(((((((((....))).))))))...((.....)))))).)))).).)))..........((((((.(((((....))))))))))).. ( -45.50) >DroSec_CAF1 2481 120 - 1 AUACCGGUCAGCAAUCGAAAAUUGAUGCGAAAUCGUUCCCAUACGGGAAGCGAUCUGGCAGCCUGCUCGCGGCUGUGAUCGAUAACAGACCGUUGCGAAUUGCGCGCGCAACCGCGACAA .....((((.(((.((((...)))))))...(((.(((((....)))))..((((..((((((.......)))))))))))))....))))((((((..((((....)))).)))))).. ( -44.40) >DroSim_CAF1 2487 120 - 1 AUACCGGCCAGCAAUCGAAAAUUGAUGCGAAAUCGUUCCCAUACGGGAAGCGAUCUGGCAGCCUGCUCGCGGCUGUGAUCGAUAACAGACCGUUGCGAAUUGCGCGCGCAACCGCGACAA ......((.(((((((((...)))))((.(.(((((((((....))).)))))).).))....)))).))((((((........)))).))((((((..((((....)))).)))))).. ( -41.90) >DroEre_CAF1 2438 120 - 1 AUACCGGCCAGCAAUCGGAAAUCGAUACAGUAUCAUUUUCAUAGAAGAGGUGAUCUGGCAGCAUGUUCGCAGCUGUGAUCGAUAACAGACCGUUGCGAGUUGCGCGCGCAACCGCCACAA .....(((..((((.(((..((((((.(((.((((((((.......)))))))))))(((((.((....))))))).))))))......)))))))(.(((((....))))))))).... ( -40.50) >DroYak_CAF1 2326 120 - 1 AUACUGGCCAGCAAUCGGAAAUCGAAACAGAAUUGUUCCCAUAAGGGGAGCAGUCUGGCAGCCUGCUCGCAGCUGUGAUCGAUAACAGACCGUUGCGAGUUGCGCGCGCAACCGCAACAA ...(((.((.......))..(((((..((((.((((((((.....))))))))))))(((((.((....)))))))..)))))..)))...((((((.(((((....))))))))))).. ( -50.00) >consensus AUACCGGCCAGCAAUCGAAAAUCGAUGCGAAAUCGUUCCCAUACGGGAAGCGAUCUGGCAGCCUGCUCGCAGCUGUGAUCGAUAACAGACCGUUGCGAGUUGCGCGCGCAACCGCGACAA .....((.(....(((((.............(((((((((....))))).))))...(((((.((....)))))))..)))))....).))((((((.(((((....))))))))))).. (-36.90 = -35.50 + -1.40)



| Location | 19,582,640 – 19,582,754 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.73 |
| Mean single sequence MFE | -33.54 |
| Consensus MFE | -19.90 |
| Energy contribution | -20.42 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19582640 114 + 22224390 GGGAACGAUUUCGCAUCGAUUUUCGAUUGCUGGCCGGUAUAAUUUGAAAAUGUUACUUU-----GCUGACCGGAUUUUGGUCCU-UGGCCAGAUGUUGGCCAUGAAUCUAAAAGCCAGUA (..((((((.....)))).))..)...(((((((((((.(((..(((.....)))..))-----)...))))..((((((....-(((((((...))))))).....))))))))))))) ( -32.20) >DroSec_CAF1 2561 118 + 1 GGGAACGAUUUCGCAUCAAUUUUCGAUUGCUGACCGGUAUAAUUUGAA-AAGUAACUUUUUCAAACUGACCGAAUUUUGGUUCU-UGGCCAGAUGUUGGCCAUGAUUCUAAAAGCUAGUA (.(((....))).).............(((((..((((....((((((-(((....)))))))))...))))..((((((.((.-(((((((...))))))).))..))))))..))))) ( -28.30) >DroSim_CAF1 2567 114 + 1 GGGAACGAUUUCGCAUCAAUUUUCGAUUGCUGGCCGGUAUAAUUUGAAAAAGUUACUUU-----GCUGACCGAAUUUUGGUUCU-UGGCCAGAUGUUGGCCAUGAUUCUAAAAGCCAGUA (.(((....))).).............((((((((((((((((((....))))))...)-----))))......((((((.((.-(((((((...))))))).))..))))))))))))) ( -32.70) >DroEre_CAF1 2518 115 + 1 GAAAAUGAUACUGUAUCGAUUUCCGAUUGCUGGCCGGUAUAAUUUGAAAAAAUUACUGC-----GCCACUCGGACUUUGAUCCUUUGGUCAGGUCCUGGUUGGGAUUCUUAAAGCCAGCA ((((.(((((...))))).))))....(((((((..(((((((((....)))))).)))-----.(((((.((((((.((((....)))))))))).)).)))..........))))))) ( -36.40) >DroYak_CAF1 2406 114 + 1 GGGAACAAUUCUGUUUCGAUUUCCGAUUGCUGGCCAGUAUAAUUUGAAAAAGUUACUGA-----GCUGCCCGGACUUUUGUCCU-UGGCCAGGUCCUGGCCAGGAUUUUUAAGGCCUGAA .((((((....))))))....((((...((.((((((((..((((....))))))))).-----))))).))))(((..(((((-.((((((...)))))))))))....)))....... ( -38.10) >consensus GGGAACGAUUUCGCAUCGAUUUUCGAUUGCUGGCCGGUAUAAUUUGAAAAAGUUACUUU_____GCUGACCGGAUUUUGGUCCU_UGGCCAGAUGUUGGCCAUGAUUCUAAAAGCCAGUA (((((((((.....)))).)))))...(((((((.....((((((....))))))...........((.((((..(((((((....)))))))..)))).))...........))))))) (-19.90 = -20.42 + 0.52)


| Location | 19,582,640 – 19,582,754 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.73 |
| Mean single sequence MFE | -27.86 |
| Consensus MFE | -16.78 |
| Energy contribution | -16.70 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19582640 114 - 22224390 UACUGGCUUUUAGAUUCAUGGCCAACAUCUGGCCA-AGGACCAAAAUCCGGUCAGC-----AAAGUAACAUUUUCAAAUUAUACCGGCCAGCAAUCGAAAAUCGAUGCGAAAUCGUUCCC ..((((((..........((((((.....))))))-..((((.......))))...-----........................))))))..((((.....))))(((....))).... ( -28.20) >DroSec_CAF1 2561 118 - 1 UACUAGCUUUUAGAAUCAUGGCCAACAUCUGGCCA-AGAACCAAAAUUCGGUCAGUUUGAAAAAGUUACUU-UUCAAAUUAUACCGGUCAGCAAUCGAAAAUUGAUGCGAAAUCGUUCCC ..................((((((.....))))))-.((((......(((((.((((((((((......))-))))))))..)))))...(((.((((...)))))))......)))).. ( -28.40) >DroSim_CAF1 2567 114 - 1 UACUGGCUUUUAGAAUCAUGGCCAACAUCUGGCCA-AGAACCAAAAUUCGGUCAGC-----AAAGUAACUUUUUCAAAUUAUACCGGCCAGCAAUCGAAAAUUGAUGCGAAAUCGUUCCC ..(((((........((.((((((.....))))))-.)).........((((....-----.....................)))))))))..(((((...)))))(((....))).... ( -22.91) >DroEre_CAF1 2518 115 - 1 UGCUGGCUUUAAGAAUCCCAACCAGGACCUGACCAAAGGAUCAAAGUCCGAGUGGC-----GCAGUAAUUUUUUCAAAUUAUACCGGCCAGCAAUCGGAAAUCGAUACAGUAUCAUUUUC (((((((..........(((....((((.(((((...)).)))..))))...)))(-----(..(((((((....)))))))..)))))))))...((((((.((((...)))))))))) ( -28.80) >DroYak_CAF1 2406 114 - 1 UUCAGGCCUUAAAAAUCCUGGCCAGGACCUGGCCA-AGGACAAAAGUCCGGGCAGC-----UCAGUAACUUUUUCAAAUUAUACUGGCCAGCAAUCGGAAAUCGAAACAGAAUUGUUCCC .................((((((((((.(((.((.-.((((....)))))).))).-----)).((((.((....)).)))).)))))))).....((((.((......))....)))). ( -31.00) >consensus UACUGGCUUUUAGAAUCAUGGCCAACAUCUGGCCA_AGGACCAAAAUCCGGUCAGC_____AAAGUAACUUUUUCAAAUUAUACCGGCCAGCAAUCGAAAAUCGAUGCGAAAUCGUUCCC ..((((((...........(((((.....)))))...(((......)))....................................))))))..(((((...))))).............. (-16.78 = -16.70 + -0.08)

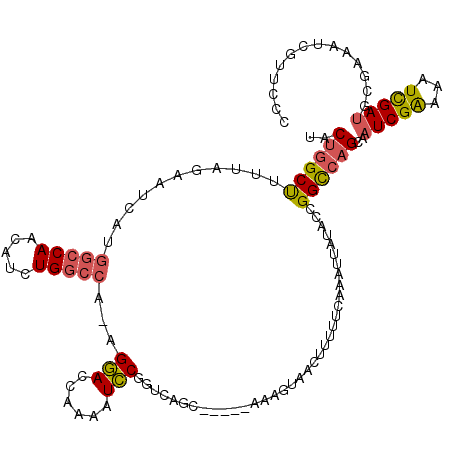
| Location | 19,582,680 – 19,582,779 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 78.30 |
| Mean single sequence MFE | -29.44 |
| Consensus MFE | -19.52 |
| Energy contribution | -17.12 |
| Covariance contribution | -2.40 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19582680 99 + 22224390 AAUUUGAAAAUGUUACUUU-----GCUGACCGGAUUUUGGUCCU-UGGCCAGAUGUUGGCCAUGAAUCUAAAAGCCAGUAUUUAGUAACUUGCGUUGAUUGCAGC ...........((((((.(-----((((...((((....)))).-(((((((...))))))).............)))))...))))))(((((.....))))). ( -27.30) >DroSec_CAF1 2601 103 + 1 AAUUUGAA-AAGUAACUUUUUCAAACUGACCGAAUUUUGGUUCU-UGGCCAGAUGUUGGCCAUGAUUCUAAAAGCUAGUAAUUAGCAACUUGCGUUGAUUGCAGC ..((((((-(((....))))))))).........((((((.((.-(((((((...))))))).))..))))))(((.(((((((((.......)))))))))))) ( -29.40) >DroSim_CAF1 2607 99 + 1 AAUUUGAAAAAGUUACUUU-----GCUGACCGAAUUUUGGUUCU-UGGCCAGAUGUUGGCCAUGAUUCUAAAAGCCAGUAAUUAGCAACUUGCGUUGAUUGCAGC (((((....))))).....-----(((....(..((((((.((.-(((((((...))))))).))..))))))..).(((((((((.......)))))))))))) ( -26.20) >DroEre_CAF1 2558 100 + 1 AAUUUGAAAAAAUUACUGC-----GCCACUCGGACUUUGAUCCUUUGGUCAGGUCCUGGUUGGGAUUCUUAAAGCCAGCAUUUAGCAACUUCUGCUGAGUGCAAC (((((....))))).....-----.(((((.((((((.((((....)))))))))).)).)))..............((((((((((.....))))))))))... ( -31.20) >DroYak_CAF1 2446 99 + 1 AAUUUGAAAAAGUUACUGA-----GCUGCCCGGACUUUUGUCCU-UGGCCAGGUCCUGGCCAGGAUUUUUAAGGCCUGAAUUUAGUAGCUUCUGCUGAGUGCAAC .........((((((((((-----.(.(((.((((....))))(-(((((((...)))))))).........)))..)...)))))))))).(((.....))).. ( -33.10) >consensus AAUUUGAAAAAGUUACUUU_____GCUGACCGGAUUUUGGUCCU_UGGCCAGAUGUUGGCCAUGAUUCUAAAAGCCAGUAUUUAGCAACUUGCGUUGAUUGCAGC ..........................((.((((..(((((((....)))))))..)))).))...............(((((((((.......)))))))))... (-19.52 = -17.12 + -2.40)

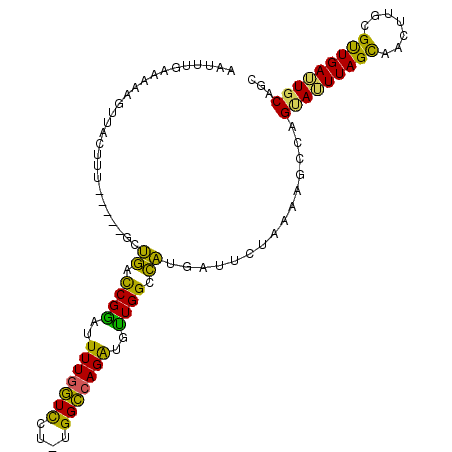

| Location | 19,582,680 – 19,582,779 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 78.30 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -15.14 |
| Energy contribution | -13.50 |
| Covariance contribution | -1.64 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19582680 99 - 22224390 GCUGCAAUCAACGCAAGUUACUAAAUACUGGCUUUUAGAUUCAUGGCCAACAUCUGGCCA-AGGACCAAAAUCCGGUCAGC-----AAAGUAACAUUUUCAAAUU ((((.....(((....))).......(((((.((((.(.(((.((((((.....))))))-.)))).)))).)))))))))-----................... ( -26.90) >DroSec_CAF1 2601 103 - 1 GCUGCAAUCAACGCAAGUUGCUAAUUACUAGCUUUUAGAAUCAUGGCCAACAUCUGGCCA-AGAACCAAAAUUCGGUCAGUUUGAAAAAGUUACUU-UUCAAAUU ((((.(((((((....))))...)))..))))........((.((((((.....))))))-.))(((.......))).((((((((((......))-)))))))) ( -25.00) >DroSim_CAF1 2607 99 - 1 GCUGCAAUCAACGCAAGUUGCUAAUUACUGGCUUUUAGAAUCAUGGCCAACAUCUGGCCA-AGAACCAAAAUUCGGUCAGC-----AAAGUAACUUUUUCAAAUU ((((....((((....))))......((((..((((.(..((.((((((.....))))))-.))..)))))..))))))))-----................... ( -25.50) >DroEre_CAF1 2558 100 - 1 GUUGCACUCAGCAGAAGUUGCUAAAUGCUGGCUUUAAGAAUCCCAACCAGGACCUGACCAAAGGAUCAAAGUCCGAGUGGC-----GCAGUAAUUUUUUCAAAUU .((((..((((..((((((((.....)).)))))).....(((......))).))))(((..((((....))))...))).-----))))............... ( -20.60) >DroYak_CAF1 2446 99 - 1 GUUGCACUCAGCAGAAGCUACUAAAUUCAGGCCUUAAAAAUCCUGGCCAGGACCUGGCCA-AGGACAAAAGUCCGGGCAGC-----UCAGUAACUUUUUCAAAUU (((((....(((....)))...........((((.........(((((((...)))))))-.((((....))))))))...-----...)))))........... ( -29.50) >consensus GCUGCAAUCAACGCAAGUUGCUAAAUACUGGCUUUUAGAAUCAUGGCCAACAUCUGGCCA_AGGACCAAAAUCCGGUCAGC_____AAAGUAACUUUUUCAAAUU ((((..((((((....))).........................(((((.....)))))...(((......))))))))))........................ (-15.14 = -13.50 + -1.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:49:24 2006