
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,570,670 – 19,570,820 |
| Length | 150 |
| Max. P | 0.996007 |

| Location | 19,570,670 – 19,570,790 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.97 |
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -18.48 |
| Energy contribution | -19.98 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19570670 120 + 22224390 CGUGUUUUUAUGUGAGCCAAGUGGUUAUGCCUGCCACCGUGCAAUGCGUUGGCUAAGUGGCUGCAAAAUGGCCAAUUUGAAGCCACCUGAAGUAGGUUGAAGUGAGUUACUGCUCACUGC .(.(((((....((.((((.(..(((((....((((.(((.....))).))))...)))))..)....))))))....))))))(((((...)))))...(((((((....))))))).. ( -38.60) >DroSec_CAF1 746 100 + 1 CGUGUUUUUAUGUGAGCCAAGUGGAUAUGCCAGCCACCGUUGAGUGCGUUGGCAAAGUGGCUGCAAAUUGGCCAAUUUGGAGCCACCUAAAGUAGGUUGA-------------------- ..............((((.........((((((((((......))).)))))))..((((((.((((((....)))))).))))))........))))..-------------------- ( -34.90) >DroSim_CAF1 686 100 + 1 CCUGUUUUUAUGUUAGCCAAGUGGAUAUGCCAGCCACCGUUGAGUGCGUUGGCUAAGUGGCUGUAAAUUGGCCAAUUUGGAGCCACCCAAAGUAGGUUGA-------------------- ............((((((...(((....(((((((((......))).))))))...((((((.((((((....)))))).))))))))).....))))))-------------------- ( -35.10) >DroEre_CAF1 724 95 + 1 -GUCUUUCUAUGAG-------U------GAUAG---CCAUUAAGUGCGGUGGCUAAGUGGCUGUAAAUUGGCCAAUUUAGAGCGCCC-UAAGUGGGUUAAAGUGAGU-------CACUAC -.............-------.------..(((---(((((......))))))))(((((((.((..((((((.((((((......)-))))).))))))..)))))-------)))).. ( -29.10) >consensus CGUGUUUUUAUGUGAGCCAAGUGGAUAUGCCAGCCACCGUUGAGUGCGUUGGCUAAGUGGCUGCAAAUUGGCCAAUUUGGAGCCACCUAAAGUAGGUUGA____________________ ......(((((.................(((((((((......))).))))))...((((((.((((((....)))))).)))))).....)))))........................ (-18.48 = -19.98 + 1.50)



| Location | 19,570,670 – 19,570,790 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.97 |
| Mean single sequence MFE | -26.67 |
| Consensus MFE | -15.29 |
| Energy contribution | -16.73 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19570670 120 - 22224390 GCAGUGAGCAGUAACUCACUUCAACCUACUUCAGGUGGCUUCAAAUUGGCCAUUUUGCAGCCACUUAGCCAACGCAUUGCACGGUGGCAGGCAUAACCACUUGGCUCACAUAAAAACACG ...((((((...............(((.....((((((((.((((........)))).)))))))).((((.((.......)).)))))))((........))))))))........... ( -34.30) >DroSec_CAF1 746 100 - 1 --------------------UCAACCUACUUUAGGUGGCUCCAAAUUGGCCAAUUUGCAGCCACUUUGCCAACGCACUCAACGGUGGCUGGCAUAUCCACUUGGCUCACAUAAAAACACG --------------------....((......((((((((.((((((....)))))).))))))))(((((.(.((((....))))).))))).........))................ ( -27.30) >DroSim_CAF1 686 100 - 1 --------------------UCAACCUACUUUGGGUGGCUCCAAAUUGGCCAAUUUACAGCCACUUAGCCAACGCACUCAACGGUGGCUGGCAUAUCCACUUGGCUAACAUAAAAACAGG --------------------....(((....(((((((((..(((((....)))))..)))))))))((((.(.((((....))))).)))).........................))) ( -25.60) >DroEre_CAF1 724 95 - 1 GUAGUG-------ACUCACUUUAACCCACUUA-GGGCGCUCUAAAUUGGCCAAUUUACAGCCACUUAGCCACCGCACUUAAUGG---CUAUC------A-------CUCAUAGAAAGAC- (.((((-------(..........(((.....-))).(((.((((((....)))))).)))....((((((..........)))---)))))------)-------)))..........- ( -19.50) >consensus ____________________UCAACCUACUUUAGGUGGCUCCAAAUUGGCCAAUUUACAGCCACUUAGCCAACGCACUCAACGGUGGCUGGCAUAUCCACUUGGCUCACAUAAAAACACG ................................((((((((.((((((....)))))).)))))))).((((.((.......)).))))................................ (-15.29 = -16.73 + 1.44)



| Location | 19,570,710 – 19,570,820 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 76.69 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -15.85 |
| Energy contribution | -18.72 |
| Covariance contribution | 2.87 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.996007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19570710 110 - 22224390 CACCCUGCUCACUUCUGGUCACUGGCCAGAGCAGUGAGCAGUAACUCACUUCAACCUACUUCAGGUGGCUUCAAAUUGGCCAUUUUGCAGCCACUUAGCCAACGCAUUGC ....(((((((((((((((.....))))))..))))))))).....................((((((((.((((........)))).)))))))).((....))..... ( -39.30) >DroSec_CAF1 786 86 - 1 CGCCCUGCUCACUUCUGGUCACUGGC------------------------UCAACCUACUUUAGGUGGCUCCAAAUUGGCCAAUUUGCAGCCACUUUGCCAACGCACUCA .((((...........))....((((------------------------............((((((((.((((((....)))))).)))))))).))))..))..... ( -21.82) >DroSim_CAF1 726 86 - 1 CACCCUGCUCACUUCUGGUCACUGGC------------------------UCAACCUACUUUGGGUGGCUCCAAAUUGGCCAAUUUACAGCCACUUAGCCAACGCACUCA .....((((((....)))....((((------------------------(...((......))((((((..(((((....)))))..))))))..)))))..))).... ( -20.00) >DroEre_CAF1 747 102 - 1 CACCCUGCUCACUUCUGGUCACUGGCCAGAGUAGUG-------ACUCACUUUAACCCACUUA-GGGCGCUCUAAAUUGGCCAAUUUACAGCCACUUAGCCACCGCACUUA .....((((((((((((((.....))))))..))))-------)..........(((.....-))).(((.((((((....)))))).)))............))).... ( -24.70) >consensus CACCCUGCUCACUUCUGGUCACUGGC________________________UCAACCUACUUUAGGUGGCUCCAAAUUGGCCAAUUUACAGCCACUUAGCCAACGCACUCA .....(((.....((((((.....))))))................................((((((((.((((((....)))))).)))))))).......))).... (-15.85 = -18.72 + 2.87)

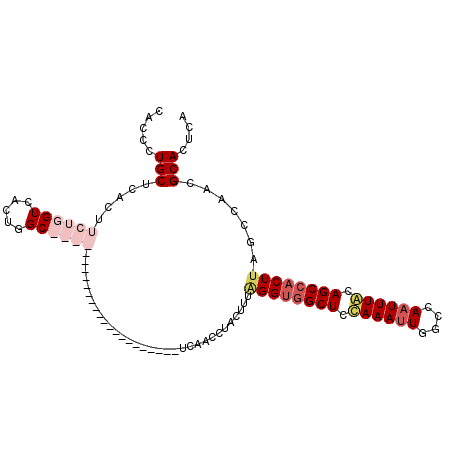

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:49:14 2006