
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,567,518 – 19,567,677 |
| Length | 159 |
| Max. P | 0.878753 |

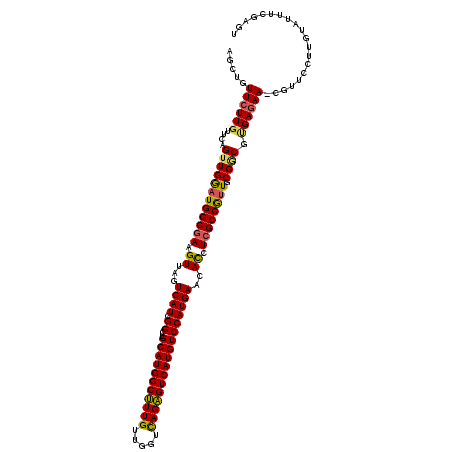
| Location | 19,567,518 – 19,567,637 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.66 |
| Mean single sequence MFE | -46.60 |
| Consensus MFE | -35.40 |
| Energy contribution | -35.18 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19567518 119 - 22224390 AGCUGUUCUUGUUCAGUUGGAUGGCGAAGUUAGUCAUCGGCUGCAUGGCUUUGUUGGUCAGAGUCAUGUCCAUGAACACCUCGCCGUUGCGGCGUGAGAA-CGUUCCUUGUAUUUCGAGU ....(((((..(...((((((((((((.((...((((.((..(((((((((((.....)))))))))))))))))..)).)))))))).)))))..))))-)....((((.....)))). ( -43.60) >DroSec_CAF1 31453 119 - 1 AGCUGUUCUUGUUCAGUUGGAUGGCGAAGUUAGUCAUCGGCUGCAUGGCUUUGUUGGUCAGAGUCAUGUCCAUGAACACUUCGCCG-UGCGGCGUGAGAACCGUUCCUUGUAUUUCGAGU (((.(((((..(...((((.((((((((((...((((.((..(((((((((((.....)))))))))))))))))..)))))))))-).)))))..))))).))).((((.....)))). ( -50.30) >DroEre_CAF1 31249 119 - 1 AGCUGUUCUUGUUCAGUUGGAUGGCGAAGUUUGUCAUCGGCUGCAUGGCUUUGUUGGUCAGAGUCAUGUCCAUGAACACCUCGCCGUUGCGGCGUGAGAA-CGUUCCUUGUAUUUCGAGU ....(((((..(...((((((((((((.((...((((.((..(((((((((((.....)))))))))))))))))..)).)))))))).)))))..))))-)....((((.....)))). ( -43.60) >DroWil_CAF1 34898 119 - 1 AACUGUUCUUGUUGAGUUGAAUGGCAAAGUUGGUCAUGGGUUGCAUGGCUUUGUUGGUUAGAGUCAUGUCCAUGAAAAUUUCGCCAUUGCGACGAGAGAA-UGUACCCUGAAUCUCAAGA .((..(((((((((.....((((((.(((((..((((((...(((((((((((.....))))))))))))))))).))))).)))))).)))))))))..-.))................ ( -41.80) >DroYak_CAF1 30137 119 - 1 AGCUGUUCUUGUUUAGUUGGAUGGCGAAGUUAGUCAUCGGCUGCAUGGCUUUGUUGGUCAGAGUCAUGUCCAUGAACACCUCGCCGUUGCGGCGUGAAAA-CGUUCCUUGUAUUUCCAGU .((((..............((((((((.((...((((.((..(((((((((((.....)))))))))))))))))..)).))))))))(..((((....)-)))..).........)))) ( -40.70) >DroAna_CAF1 37088 119 - 1 AGCUGUUCUUGUUCAGCUGGAUGGCGAAGUUGGUCAUCGGCUGCAUGGCCUUGUUGGUCAGGGUCAUGUCCAUGAACACUUCGCCGUUGCGCCGCGAGAA-GGUGCCCUGGAUCUCCAGG (((((........))))).(((((((((((...((((.((..(((((((((((.....)))))))))))))))))..)))))))))))(((((.......-)))))(((((....))))) ( -59.60) >consensus AGCUGUUCUUGUUCAGUUGGAUGGCGAAGUUAGUCAUCGGCUGCAUGGCUUUGUUGGUCAGAGUCAUGUCCAUGAACACCUCGCCGUUGCGGCGUGAGAA_CGUUCCUUGUAUUUCGAGU .....((((((....((((((((((((.((...((((.((..(((((((((((.....)))))))))))))))))..)).)))))))).)))).)))))).................... (-35.40 = -35.18 + -0.22)


| Location | 19,567,557 – 19,567,677 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -46.40 |
| Consensus MFE | -41.51 |
| Energy contribution | -41.32 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.878753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19567557 120 - 22224390 AGGGGAGCAGCCUGCAUGGGCGACGCGGGCACCAGGCCGAAGCUGUUCUUGUUCAGUUGGAUGGCGAAGUUAGUCAUCGGCUGCAUGGCUUUGUUGGUCAGAGUCAUGUCCAUGAACACC .((((((((((..((.(((((.......)).))).))....))))))))((((((....((((((.......))))))((..(((((((((((.....))))))))))))).)))))))) ( -48.50) >DroSec_CAF1 31492 120 - 1 AGGGGAGCAGCCUGCAUGGGCGACGCGGGCACCAGGCCGAAGCUGUUCUUGUUCAGUUGGAUGGCGAAGUUAGUCAUCGGCUGCAUGGCUUUGUUGGUCAGAGUCAUGUCCAUGAACACU ..(((((((((..((.(((((.......)).))).))....)))))))))(((((....((((((.......))))))((..(((((((((((.....))))))))))))).)))))... ( -48.30) >DroEre_CAF1 31288 120 - 1 AGGGGAGCAGCCUGCAUGGGCGACGCGGGUACCAGGCCGAAGCUGUUCUUGUUCAGUUGGAUGGCGAAGUUUGUCAUCGGCUGCAUGGCUUUGUUGGUCAGAGUCAUGUCCAUGAACACC .((((((((((..((.(((...((....)).))).))....))))))))((((((....((((((((...))))))))((..(((((((((((.....))))))))))))).)))))))) ( -47.10) >DroWil_CAF1 34937 120 - 1 AGAGGAGUAGCCUGCAAUGGCGCCGAUGGCGAUAAGCCGAAACUGUUCUUGUUGAGUUGAAUGGCAAAGUUGGUCAUGGGUUGCAUGGCUUUGUUGGUUAGAGUCAUGUCCAUGAAAAUU .........(((......)))((((.((((.....))))...(..(((.....)))..)..))))..((((..((((((...(((((((((((.....))))))))))))))))).)))) ( -37.40) >DroYak_CAF1 30176 120 - 1 AGCGGAGCAGCCUGCAUGGGCGACGCGGGCACCAGGCCGAAGCUGUUCUUGUUUAGUUGGAUGGCGAAGUUAGUCAUCGGCUGCAUGGCUUUGUUGGUCAGAGUCAUGUCCAUGAACACC ...((((((((..((.(((((.......)).))).))....))))))))((((((....((((((.......))))))((..(((((((((((.....))))))))))))).)))))).. ( -45.50) >DroAna_CAF1 37127 120 - 1 AGUGGAGCAGCCUGCAGCGGCGACGAUGGCACCAGGCCGAAGCUGUUCUUGUUCAGCUGGAUGGCGAAGUUGGUCAUCGGCUGCAUGGCCUUGUUGGUCAGGGUCAUGUCCAUGAACACU .((((((((((((((..((....))...)))...((((((.(((((((..........)))))))....))))))...))))))(((((((((.....))))))))).)))))....... ( -51.60) >consensus AGGGGAGCAGCCUGCAUGGGCGACGCGGGCACCAGGCCGAAGCUGUUCUUGUUCAGUUGGAUGGCGAAGUUAGUCAUCGGCUGCAUGGCUUUGUUGGUCAGAGUCAUGUCCAUGAACACC ((((.(((.(((......)))......(((.....)))...))).))))((((((....((((((.......))))))((..(((((((((((.....))))))))))))).)))))).. (-41.51 = -41.32 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:49:10 2006