| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,565,346 – 19,565,466 |
| Length | 120 |
| Max. P | 0.792280 |

| Location | 19,565,346 – 19,565,466 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.06 |
| Mean single sequence MFE | -31.45 |
| Consensus MFE | -29.46 |
| Energy contribution | -29.77 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.571080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19565346 120 + 22224390 CCAAGUACUUCACCACCACCAAGAAGGGCGAGAUCUUCGAGCUCAAGUCGGAGCUGAAUAACGACAAGAAGGAGAAGAAGAAGGAGGCGGUGAAGAAGGUGAUCGCCAGCAUGACCGUGG (((....(((((((.((.((..(((((......))))).(((((......)))))...........................)).)).)))))))..(((.(((....).)).))).))) ( -34.80) >DroSec_CAF1 29234 120 + 1 CCAAGUACUUCACCACCACCAAGAAGGGCGAGAUCUUCGAGCUCAAGUCGGAGCUGAAUAACGACAAGAAGGAGAAGAAGAAGGAGGCGGUGAAGAAGGUGAUCGCCAGCAUGACCGUGG (((....(((((((.((.((..(((((......))))).(((((......)))))...........................)).)).)))))))..(((.(((....).)).))).))) ( -34.80) >DroEre_CAF1 29079 120 + 1 CCAAGUACUUCACCACCACCAAGAAGGGCGAGAUCUUCGAGCUGAAGUCGGAGCUUAACAAUGACAAGAAGGAGAAAAAGAAGGAGGCGGUGAAGAAGGUGAUCGCUAGCAUGACCGUGG (((....(((((((.((.((..(((((......)))))(((((........)))))..........................)).)).)))))))..(((.(((....).)).))).))) ( -32.70) >DroWil_CAF1 32714 120 + 1 CCAAAUAUUUCACCACCACCAAGAAGGGUGAAAUCUUCGAGCUUAAGUCGGAGCUAAACAAUGAUAAGAAGGAGAAGAAGAAGGAGGCGGUAAAGAAGGUGAUUGCCAGCAUGACUGUGG ((.....(((((((............)))))))(((((...(((...((....(........)....))..)))..))))).))..(((((...(..(((....)))..)...))))).. ( -25.20) >DroYak_CAF1 27967 120 + 1 CCAAGUACUUCACCACCACCAAGAAGGGCGAGAUCUUCGAGCUGAAGUCGGAGCUGAACAAUGACAAGAAGGAGAAAAAGAAGGAGGCGGUGAAGAAGGUGAUCGCCAGCAUGACCGUGG (((....(((((((.((.((..(((((......))))).((((........))))...........................)).)).)))))))..(((.(((....).)).))).))) ( -31.50) >DroAna_CAF1 34913 120 + 1 CCAAGUACUUCACCACCACCAAGAAGGGCGAGAUCUUCGAGCUCAAAUCGGAGCUGAACAAUGACAAGAAGGAGAAGAAGAAGGAGGCCGUGAAGAAGGUGAUAGCCAGCAUGACGGUGG (((.((.((((((..((.((..(((((......))))).(((((......)))))...........................)).))..))))))..(((....)))......))..))) ( -29.70) >consensus CCAAGUACUUCACCACCACCAAGAAGGGCGAGAUCUUCGAGCUCAAGUCGGAGCUGAACAAUGACAAGAAGGAGAAGAAGAAGGAGGCGGUGAAGAAGGUGAUCGCCAGCAUGACCGUGG (((....(((((((.((.((..(((((......))))).(((((......)))))...........................)).)).)))))))..(((....)))..........))) (-29.46 = -29.77 + 0.31)

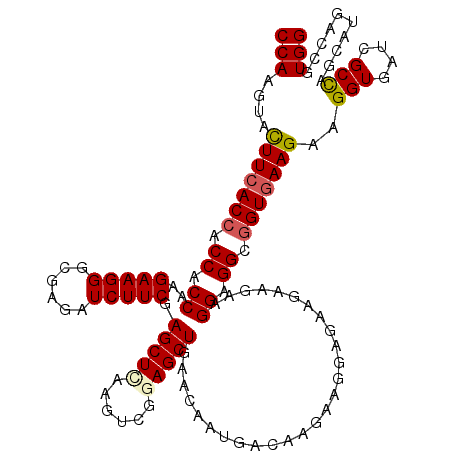
| Location | 19,565,346 – 19,565,466 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.06 |
| Mean single sequence MFE | -30.41 |
| Consensus MFE | -26.74 |
| Energy contribution | -27.10 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
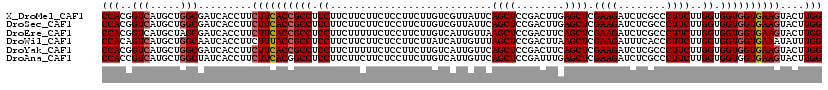
>X_DroMel_CAF1 19565346 120 - 22224390 CCACGGUCAUGCUGGCGAUCACCUUCUUCACCGCCUCCUUCUUCUUCUCCUUCUUGUCGUUAUUCAGCUCCGACUUGAGCUCGAAGAUCUCGCCCUUCUUGGUGGUGGUGAAGUACUUGG (((.((((........)))).....((((((((((.......(((((...(((..((((...........))))..)))...)))))....(((......)))))))))))))....))) ( -33.60) >DroSec_CAF1 29234 120 - 1 CCACGGUCAUGCUGGCGAUCACCUUCUUCACCGCCUCCUUCUUCUUCUCCUUCUUGUCGUUAUUCAGCUCCGACUUGAGCUCGAAGAUCUCGCCCUUCUUGGUGGUGGUGAAGUACUUGG (((.((((........)))).....((((((((((.......(((((...(((..((((...........))))..)))...)))))....(((......)))))))))))))....))) ( -33.60) >DroEre_CAF1 29079 120 - 1 CCACGGUCAUGCUAGCGAUCACCUUCUUCACCGCCUCCUUCUUUUUCUCCUUCUUGUCAUUGUUAAGCUCCGACUUCAGCUCGAAGAUCUCGCCCUUCUUGGUGGUGGUGAAGUACUUGG (((.((((........)))).....((((((((((.((...........................((((........)))).((((........))))..)).))))))))))....))) ( -30.10) >DroWil_CAF1 32714 120 - 1 CCACAGUCAUGCUGGCAAUCACCUUCUUUACCGCCUCCUUCUUCUUCUCCUUCUUAUCAUUGUUUAGCUCCGACUUAAGCUCGAAGAUUUCACCCUUCUUGGUGGUGGUGAAAUAUUUGG ((((((.....)))............(((((((((.((...........................((((........)))).((((........))))..)).))))))))).....))) ( -24.90) >DroYak_CAF1 27967 120 - 1 CCACGGUCAUGCUGGCGAUCACCUUCUUCACCGCCUCCUUCUUUUUCUCCUUCUUGUCAUUGUUCAGCUCCGACUUCAGCUCGAAGAUCUCGCCCUUCUUGGUGGUGGUGAAGUACUUGG (((.((((........)))).....((((((((((.((...........................((((........)))).((((........))))..)).))))))))))....))) ( -30.10) >DroAna_CAF1 34913 120 - 1 CCACCGUCAUGCUGGCUAUCACCUUCUUCACGGCCUCCUUCUUCUUCUCCUUCUUGUCAUUGUUCAGCUCCGAUUUGAGCUCGAAGAUCUCGCCCUUCUUGGUGGUGGUGAAGUACUUGG ...(((...((((.(((((((((........(((.....(((((..........((........))((((......))))..)))))....)))......)))))))))..))))..))) ( -30.14) >consensus CCACGGUCAUGCUGGCGAUCACCUUCUUCACCGCCUCCUUCUUCUUCUCCUUCUUGUCAUUGUUCAGCUCCGACUUGAGCUCGAAGAUCUCGCCCUUCUUGGUGGUGGUGAAGUACUUGG (((..(((.....))).........((((((((((.((...........................((((........)))).((((........))))..)).))))))))))....))) (-26.74 = -27.10 + 0.36)
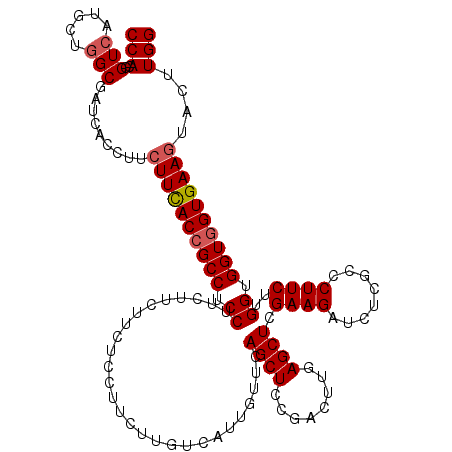
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:49:08 2006