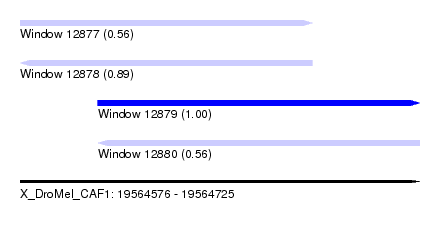
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,564,576 – 19,564,725 |
| Length | 149 |
| Max. P | 0.999988 |

| Location | 19,564,576 – 19,564,685 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 69.62 |
| Mean single sequence MFE | -21.87 |
| Consensus MFE | -9.80 |
| Energy contribution | -10.99 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19564576 109 + 22224390 ----------AGCCUGUUGCUUUUACGAUCCGUUACUUUUCAGAUGUAAUAUAGCCAUCUCACUCUAAUUCGCUAUAUUGCUCUCUGGUGAGAGCGUCGACUUCGUUUUUCAUUGAGAU ----------(((.....)))....(((..((((.(((.(((((.((((((((((................))))))))))..))))).))))))))))..((((........)))).. ( -23.69) >DroSec_CAF1 28484 93 + 1 ----------AGCCUGUUGCUUUUUCGUUCCGCU-UUUUUCAGAUGUAAUAUAGUCAUCUCACUCUAAUUCGCUAUAUUGCUCUCUAGUGAGAGCGUCGACUUC--------------- ----------(((.....)))...(((...((((-(((...(((.((((((((((................))))))))))..)))...))))))).)))....--------------- ( -20.59) >DroEre_CAF1 28297 117 + 1 AUUUCUUUGAAGUUCGUUGCUUUUACGAUCCGUUACUUUUCAGUUUUCAUAUAGUCAUCUCACUCUAAUUCGCCAAAUUGCUCUAUCGUGAGAGCGUCUACUUAC--UUUCAUUGAGAU (((((..((((((..((((((((((((((..(..(((....)))...).......................((......))...))))))))))))...))..))--.))))..))))) ( -21.40) >DroYak_CAF1 27232 90 + 1 ---------------------------GUCCGUUACUUUUAAGUUGUAAUAUAGUCAUCUCGCUCUAAUUCAGUAAGUUGCUCUCUUGUGAGAGCGUUAACUUAC--UUUCAUUGAGAU ---------------------------....(((((.........)))))............(((......((((((((((((((....))))))...)))))))--)......))).. ( -21.80) >consensus __________AGCCUGUUGCUUUUACGAUCCGUUACUUUUCAGAUGUAAUAUAGUCAUCUCACUCUAAUUCGCUAAAUUGCUCUCUAGUGAGAGCGUCGACUUAC__UUUCAUUGAGAU ..............................((((.(((.((((..((((((((((................))))))))))...)))).)))))))....................... ( -9.80 = -10.99 + 1.19)


| Location | 19,564,576 – 19,564,685 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 69.62 |
| Mean single sequence MFE | -21.98 |
| Consensus MFE | -10.99 |
| Energy contribution | -11.05 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19564576 109 - 22224390 AUCUCAAUGAAAAACGAAGUCGACGCUCUCACCAGAGAGCAAUAUAGCGAAUUAGAGUGAGAUGGCUAUAUUACAUCUGAAAAGUAACGGAUCGUAAAAGCAACAGGCU---------- ((((((.(((....((....))..((((((....))))))...........)))...)))))).(((...(((((((((........))))).)))).)))........---------- ( -25.80) >DroSec_CAF1 28484 93 - 1 ---------------GAAGUCGACGCUCUCACUAGAGAGCAAUAUAGCGAAUUAGAGUGAGAUGACUAUAUUACAUCUGAAAAA-AGCGGAACGAAAAAGCAACAGGCU---------- ---------------....(((..((((((....))))))......((...(((((((((..........)))).)))))....-.))....)))...(((.....)))---------- ( -20.30) >DroEre_CAF1 28297 117 - 1 AUCUCAAUGAAA--GUAAGUAGACGCUCUCACGAUAGAGCAAUUUGGCGAAUUAGAGUGAGAUGACUAUAUGAAAACUGAAAAGUAACGGAUCGUAAAAGCAACGAACUUCAAAGAAAU .(((...((((.--((..((((.((.((((((..(((.((......))...)))..)))))))).))))......(((....)))......((((.......)))))))))).)))... ( -20.60) >DroYak_CAF1 27232 90 - 1 AUCUCAAUGAAA--GUAAGUUAACGCUCUCACAAGAGAGCAACUUACUGAAUUAGAGCGAGAUGACUAUAUUACAACUUAAAAGUAACGGAC--------------------------- (((((..(((.(--(((((((...((((((....))))))))))))))...)))....))))).............................--------------------------- ( -21.20) >consensus AUCUCAAUGAAA__GGAAGUCGACGCUCUCACAAGAGAGCAAUAUAGCGAAUUAGAGUGAGAUGACUAUAUUACAACUGAAAAGUAACGGAUCGUAAAAGCAACAGGCU__________ .................(((((.(((((((....))))))......((........))..).))))).................................................... (-10.99 = -11.05 + 0.06)

| Location | 19,564,605 – 19,564,725 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.11 |
| Mean single sequence MFE | -26.67 |
| Consensus MFE | -17.93 |
| Energy contribution | -20.24 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.67 |
| SVM decision value | 5.48 |
| SVM RNA-class probability | 0.999988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19564605 120 + 22224390 UCAGAUGUAAUAUAGCCAUCUCACUCUAAUUCGCUAUAUUGCUCUCUGGUGAGAGCGUCGACUUCGUUUUUCAUUGAGAUUUCAGUUUACUUCAUUGGCUGUCCUUUAUUUUAUAUCUGG .(((((((((.((((((((((((....((..((......(((((((....))))))).......))..))....)))))....((....))....)))))))........))))))))). ( -31.52) >DroSec_CAF1 28512 97 + 1 UCAGAUGUAAUAUAGUCAUCUCACUCUAAUUCGCUAUAUUGCUCUCUAGUGAGAGCGUCGACUUC-----------------------ACUUCAUCGGCUGUCCUUUAUAUUAUAUCUGG .((((((((((((((.........................((((((....))))))(((((....-----------------------......)))))......)))))))))))))). ( -29.20) >DroEre_CAF1 28336 107 + 1 UCAGUUUUCAUAUAGUCAUCUCACUCUAAUUCGCCAAAUUGCUCUAUCGUGAGAGCGUCUACUUAC--UUUCAUUGAGAUAUCAGUU---UUCAUCGGCUGUC-UUUAUGUGA------- .......((((((((..((((((.................(((((......)))))((......))--......))))))..(((((---......)))))..-.))))))))------- ( -19.30) >consensus UCAGAUGUAAUAUAGUCAUCUCACUCUAAUUCGCUAUAUUGCUCUCUAGUGAGAGCGUCGACUUC___UUUCAUUGAGAU_UCAGUU_ACUUCAUCGGCUGUCCUUUAUAUUAUAUCUGG .((((((((((((((((.......................((((((....))))))................(((((....)))))..........)))))).......)))))))))). (-17.93 = -20.24 + 2.31)


| Location | 19,564,605 – 19,564,725 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.11 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -12.52 |
| Energy contribution | -13.30 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19564605 120 - 22224390 CCAGAUAUAAAAUAAAGGACAGCCAAUGAAGUAAACUGAAAUCUCAAUGAAAAACGAAGUCGACGCUCUCACCAGAGAGCAAUAUAGCGAAUUAGAGUGAGAUGGCUAUAUUACAUCUGA .(((((..........((....))......((((...(..((((((.(((....((....))..((((((....))))))...........)))...))))))..)....))))))))). ( -27.10) >DroSec_CAF1 28512 97 - 1 CCAGAUAUAAUAUAAAGGACAGCCGAUGAAGU-----------------------GAAGUCGACGCUCUCACUAGAGAGCAAUAUAGCGAAUUAGAGUGAGAUGACUAUAUUACAUCUGA .(((((.(((((((..((....))........-----------------------...((((.(((((((....))))))......((........))..).))))))))))).))))). ( -24.60) >DroEre_CAF1 28336 107 - 1 -------UCACAUAAA-GACAGCCGAUGAA---AACUGAUAUCUCAAUGAAA--GUAAGUAGACGCUCUCACGAUAGAGCAAUUUGGCGAAUUAGAGUGAGAUGACUAUAUGAAAACUGA -------...((((.(-(.(((........---..))).(((((((.(((..--((......))(((((......)))))...........)))...))))))).)).))))........ ( -17.90) >consensus CCAGAUAUAAAAUAAAGGACAGCCGAUGAAGU_AACUGA_AUCUCAAUGAAA___GAAGUCGACGCUCUCACCAGAGAGCAAUAUAGCGAAUUAGAGUGAGAUGACUAUAUUACAUCUGA ...................(((..........................................((((((....))))))(((((((((..((.....))..)).)))))))....))). (-12.52 = -13.30 + 0.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:49:06 2006