
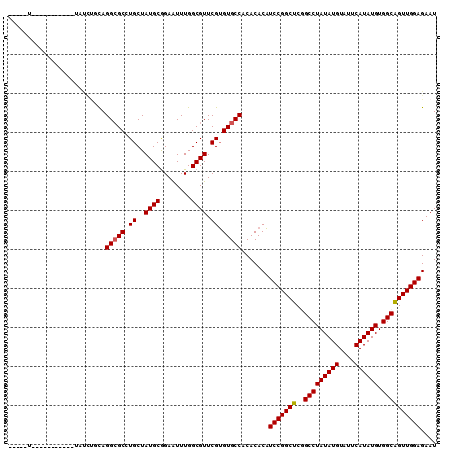
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,557,195 – 19,557,358 |
| Length | 163 |
| Max. P | 0.899518 |

| Location | 19,557,195 – 19,557,303 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.21 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -28.95 |
| Energy contribution | -28.90 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19557195 108 + 22224390 --CUAUAUUAAUUCAUUGAUCCACAGGCGCCUGCUAUGCAGAGUUUGGCGUUCGUGUGCCACACACAUCCGGCUCGGCCUAUAUGUAUUCAUAUGUGGCGGUUGGAGAAU --........((((.(..((((((..(((((.(((......)))..)))))..))).(((((........(((...))).(((((....)))))))))))))..).)))) ( -34.60) >DroVir_CAF1 28576 94 + 1 ----------------AUUUGUACAGGCGCGUGCUAUGCGGAAUUUGGCGUUCGUGUGCCCCACACAUCCGGCUCGGCCUAUAUGUAUUCAUAUGUGGCAGUUGGAGAAU ----------------...(((...((((((((..((((........)))).))))))))..)))..(((((((..(((((((((....)))))).)))))))))).... ( -30.70) >DroGri_CAF1 24618 93 + 1 -----------------UAUUUGCAGGCGCGUGCUAUGCGGAAUUUGGCGUUCGUGUGCCACACACAUCCGGCUCUGCCUAUAUGUAUUCAUAUGUGGCAGUUGGAGAAU -----------------........((((((((..((((........)))).)))))))).......(((((((..(((((((((....)))))).)))))))))).... ( -31.10) >DroWil_CAF1 24506 105 + 1 -----UAUCAUUUCAAUCUCCAUUAGGCGCUUGCUAUGCGGAAUUUGGCGUUCGUGUGCCACACACAUCCGGCUCGGCCUAUAUGUAUUCAUAUGUGGCAGUUGGAGAAU -----...........((((((....(((..(((((.........)))))..))).((((((........(((...))).(((((....)))))))))))..)))))).. ( -31.00) >DroMoj_CAF1 27128 100 + 1 CUGUAU----------AUCCUUGUAGGCGCUUGCUAUGCGGAAUUUGGCGUUCGAGUGCCACACACUUCCGGCUCAGCCUAUAUGUAUUCAUAUGUGGCAGUUGGAGAAU ......----------.....(((.((((((((..((((........)))).)))))))))))...((((((((..(((((((((....)))))).)))))))))))... ( -35.70) >DroAna_CAF1 26513 104 + 1 CUGAAUAUCAACUC------CUGCAGGAGCCUGCUAUGCGGAGUUUGGCGUUCGUGUGCCACACACAUCCGGCUCGGCCUAUAUGUAUUCAUAUGUGGCGGUUGGAGAAU ..((((..((((((------(.((((....)))).....)))).)))..))))(((((...))))).(((((((..(((((((((....)))))).)))))))))).... ( -39.70) >consensus _____U___________UAUCUGCAGGCGCCUGCUAUGCGGAAUUUGGCGUUCGUGUGCCACACACAUCCGGCUCGGCCUAUAUGUAUUCAUAUGUGGCAGUUGGAGAAU .........................(((((.((..((((........)))).)).))))).......(((((((..(((((((((....)))))).)))))))))).... (-28.95 = -28.90 + -0.05)


| Location | 19,557,265 – 19,557,358 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 94.82 |
| Mean single sequence MFE | -10.73 |
| Consensus MFE | -9.68 |
| Energy contribution | -9.43 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19557265 93 - 22224390 ---CACUCACCUAUUAGAUAUUCCAAUAUCAUGUUCCAACCAAUUAUAAAAGCUACAAAUUCUCCAACCGCCACAUAUGAAUACAUAUAGGCCGAG ---..(((........(((((....))))).......................................(((..(((((....))))).))).))) ( -10.80) >DroVir_CAF1 28632 93 - 1 ---GUCUUACCUAUUAGAUAUUCCAAUAUCAUGUUCCAACCAAUUAUAAAAGCUACAAAUUCUCCAACUGCCACAUAUGAAUACAUAUAGGCCGAG ---.............(((((....))))).....................................(.(((..(((((....))))).))).).. ( -8.50) >DroGri_CAF1 24673 96 - 1 GAUUGCUUACCUAUUAGAUAUUCCAAUAUCAUGUUCCAACCAAUUAUAAAAGCUACAAAUUCUCCAACUGCCACAUAUGAAUACAUAUAGGCAGAG ....((((........(((((....))))).((.......)).......))))..............(((((..(((((....))))).))))).. ( -14.40) >DroSec_CAF1 21191 93 - 1 ---CACUCACCUAUUAGAUAUUCCAAUAUCAUAUUCCAACCAAUUAUAAAAGCUACGAAUUCUCCAACCGCCACAUAUGAAUACAUAUAGGCCGAG ---..(((........(((((....))))).......................................(((..(((((....))))).))).))) ( -10.80) >DroMoj_CAF1 27190 93 - 1 ---CACUUACCUAUUAGAUAUUCCAAUAUCAUGUUCCAACCAAUUAUAAAAGCUACAAAUUCUCCAACUGCCACAUAUGAAUACAUAUAGGCUGAG ---..(((((((((..(((((....))))).............(((((...((................))....)))))......))))).)))) ( -9.09) >DroAna_CAF1 26579 93 - 1 ---AACUCACCUAUUAGAUAUUCCAAUAUCAUGUUCCAACCAAUUAUAAAAGCUACAAAUUCUCCAACCGCCACAUAUGAAUACAUAUAGGCCGAG ---..(((........(((((....))))).......................................(((..(((((....))))).))).))) ( -10.80) >consensus ___CACUCACCUAUUAGAUAUUCCAAUAUCAUGUUCCAACCAAUUAUAAAAGCUACAAAUUCUCCAACCGCCACAUAUGAAUACAUAUAGGCCGAG .....(((........(((((....))))).......................................(((..(((((....))))).))).))) ( -9.68 = -9.43 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:57 2006